bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
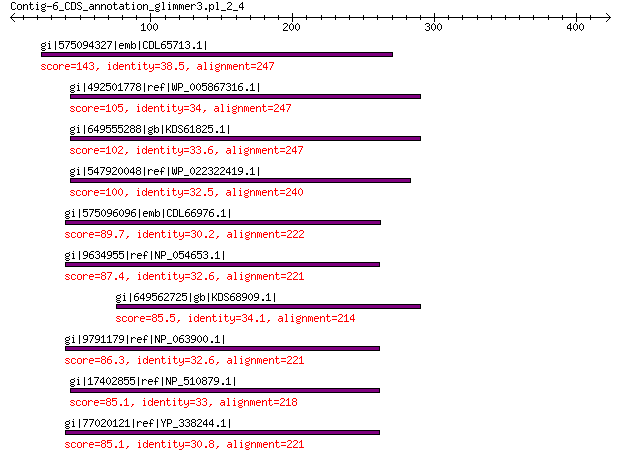
Query= Contig-6_CDS_annotation_glimmer3.pl_2_4
Length=423
Score E
Sequences producing significant alignments: (Bits) Value
gi|575094327|emb|CDL65713.1| unnamed protein product 143 2e-34
gi|492501778|ref|WP_005867316.1| hypothetical protein 105 1e-22
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 102 1e-21
gi|547920048|ref|WP_022322419.1| putative replication protein 100 1e-20
gi|575096096|emb|CDL66976.1| unnamed protein product 89.7 7e-17
gi|9634955|ref|NP_054653.1| nonstructural protein 87.4 7e-16
gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 85.5 1e-15
gi|9791179|ref|NP_063900.1| conserved hypothetical protein 86.3 2e-15
gi|17402855|ref|NP_510879.1| hypothetical protein PhiCPG1p9 85.1 2e-15
gi|77020121|ref|YP_338244.1| putative replication protein 85.1 4e-15
>gi|575094327|emb|CDL65713.1| unnamed protein product [uncultured bacterium]
Length=515
Score = 143 bits (360), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 95/296 (32%), Positives = 141/296 (48%), Gaps = 49/296 (17%)
Query 23 IDKYTIVNPATGETFPMFLIVPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYN 82
I+ ++N TG+ P+++ VPC C +C ++KA + RA+ E+ + FITLTYN
Sbjct 59 IEHCYLLNSETGDMIPLYIAVPCGSCIICRKRKANALATRAIMETEITGSSPLFITLTYN 118
Query 83 NEHLPKN-----GVFPEEIQLFFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIIL 137
EHLPKN + ++QLFFKRLR+ LD + I H+LRY+A EYG +KRPHYH++L
Sbjct 119 PEHLPKNQYGYLTLRKLDLQLFFKRLRSLLDNQSIPHSLRYLACGEYGSNTKRPHYHLLL 178
Query 138 WNFPDNF-----------ESAYS---------RLTLIESCWRRPTGEY------------ 165
W FP ++ + A+S R+ C P +Y
Sbjct 179 WGFPLSYFKDILKAQAFIQKAWSYFQVDENGKRIPYYSKCRTCPFNQYKDRRSCSDVAHF 238
Query 166 -------HPDGSPVTRS--IGFAYCVPVINGGINYVMKYMGKRECAP-EGMNSTFMLASR 215
+P G+ + R IG +P +G Y+ KYM K AP F AS
Sbjct 239 CTGVRLRYPSGAFIYRRYPIGSIKVLPANSGAPAYITKYMVKGSNAPHSSCEPPFRTASN 298
Query 216 KNGGIGSAYAEQLRAFYEQQPDTCDMSVINIYTGQPLTTMLP--RYYRMKFMPSTS 269
+ GGIGSAY + + P + V++ TG +P + + +PS S
Sbjct 299 RGGGIGSAYIRAHKDEILRNPSLEALPVVDRVTGSGKLFYMPIDSWVKSTLIPSPS 354
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 105 bits (263), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 84/265 (32%), Positives = 115/265 (43%), Gaps = 56/265 (21%)
Query 43 VPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPK------------NG 90
VPC +C C + K Q W +R E+ + F+TLTY++EH+P
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEA-DEYPFSLFVTLTYDDEHMPTAMIGEDLFKSTVGV 73
Query 91 VFPEEIQLFFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWNFPDNFESAYSR 150
V +IQLF KRLR K D+ + LRY SEYG RPHYH+IL+ FP F +
Sbjct 74 VSKRDIQLFMKRLRKKYDQ----YRLRYFLTSEYGSQGGRPHYHMILFGFP--FTGKHGG 127
Query 151 LTLIESCWRRPTGEYHPDGSPVTRSIGFAYCVPVINGGINYVMKYMGKRECAPEGMNST- 209
L+ CW+ GF P+ I YV KYM ++ P+ +
Sbjct 128 -DLLAECWKN----------------GFVQAHPLTTKEIAYVTKYMYEKSMVPDILKDVK 170
Query 210 ----FMLASRKNGGIGSAYA-EQLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYYRMKF 264
FML SR GIG + EQ+ FY P + + G + +PRYY K
Sbjct 171 EYQPFMLCSRIP-GIGYHFLREQILDFYRLHP----RDYVRAFNG--MRMAMPRYYADKL 223
Query 265 MPSTSMCYDSNFIKYFKDTVRWFEI 289
YD + +Y K+ F I
Sbjct 224 -------YDDDMKEYLKELREAFFI 241
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 102 bits (255), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 83/265 (31%), Positives = 114/265 (43%), Gaps = 56/265 (21%)
Query 43 VPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPK------------NG 90
VPC +C C + K Q W +R E+ + F+TLTY++EH+P
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEA-DEYPFSLFVTLTYDDEHIPTAMIGEDLFKTTVGV 73
Query 91 VFPEEIQLFFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWNFPDNFESAYSR 150
V +IQLF KRLR K + LRY SEYG RPHYH+IL+ FP F +
Sbjct 74 VSKRDIQLFMKRLRKKY----AQYRLRYFLTSEYGSQGGRPHYHMILFGFP--FTGKHGG 127
Query 151 LTLIESCWRRPTGEYHPDGSPVTRSIGFAYCVPVINGGINYVMKYMGKRECAPEGMNST- 209
L+ CW+ GF P+ I+YV KYM ++ P+ +
Sbjct 128 -DLLAECWKN----------------GFVQAHPLTTKEISYVTKYMYEKSMIPDILKGVK 170
Query 210 ----FMLASRKNGGIGSAYA-EQLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYYRMKF 264
FML S K GIG + EQ+ FY P + + G + +PRYY K
Sbjct 171 EYQPFMLCS-KMPGIGYHFLREQILDFYRLHP----RDYVRAFNG--MRMAMPRYYADKL 223
Query 265 MPSTSMCYDSNFIKYFKDTVRWFEI 289
YD + +Y K+ F I
Sbjct 224 -------YDDDMKEYLKELREAFFI 241
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 100 bits (249), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 78/258 (30%), Positives = 113/258 (44%), Gaps = 56/258 (22%)
Query 43 VPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVFPE-------- 94
VPC C C + K Q W +R E+ + F+TLTY++EHLP + +
Sbjct 10 VPCGWCVNCRQNKRQSWVYRLQAEA-KEYPLSLFVTLTYDDEHLPIERIGSDLFQTNVAV 68
Query 95 ----EIQLFFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWNFPDNFESAYSR 150
++QLF KRLR K + + +RY SEYG + RPHYH+IL+ FP + A
Sbjct 69 VSKRDVQLFMKRLRKKYE----DYKMRYFVTSEYGAKNGRPHYHMILFGFPFTGKMAGD- 123
Query 151 LTLIESCWRRPTGEYHPDGSPVTRSIGFAYCVPVINGGINYVMKYMGKRECAPEGMNST- 209
L+ CW+ GF P+ I YV KYM ++ PE +
Sbjct 124 --LLAECWQN----------------GFVQAHPLTIKEIAYVCKYMYEKSMCPEILRDEK 165
Query 210 ----FMLASRKNGGIGSAYAE-QLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYYRMKF 264
FML SR N GIG + + + FY + P + + G + +PRYY K
Sbjct 166 KYKPFMLCSR-NPGIGFGFMKADIIEFYRRHP----RDYVRAWAGHKMA--MPRYYADKL 218
Query 265 MPSTSMCYDSNFIKYFKD 282
YD + + K+
Sbjct 219 -------YDDDMKAFLKE 229
>gi|575096096|emb|CDL66976.1| unnamed protein product [uncultured bacterium]
Length=296
Score = 89.7 bits (221), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 67/230 (29%), Positives = 103/230 (45%), Gaps = 25/230 (11%)
Query 40 FLIVPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNGVF-PEEIQL 98
+++VPC +C C +A +W+ R C S+ + F+TLTYN+++LP NG + +Q
Sbjct 33 YVLVPCGQCLECRLHRASEWALRC-CHELKSHDKGIFLTLTYNDDNLPPNGTLVKKHVQD 91
Query 99 FFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWNFPDNFESAYSRLTLI--ES 156
F KRLR +D G +RY+ EYG S RPHYH++++ + + L I S
Sbjct 92 FIKRLRRHIDYYGDCTKIRYLCAGEYGDLSLRPHYHLLVFGYYPSDPRLLHGLQKIGKNS 151
Query 157 CWRRPT-----GEYHPDGSPVTRSIGFAYCVPVINGGINYVMKYMGKRECAPEGMNSTFM 211
+ PT G+ H +T C + Y R PE FM
Sbjct 152 LFTSPTLTKLWGKGHISFGAITFESARYTCQYALKKQTGEHSHYYVDRGVIPE-----FM 206
Query 212 LASRKNGGIGSAYAEQLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYYR 261
+ S +N G+G +A +E+ T + I I PRYY+
Sbjct 207 ICSNRN-GLGYDFAVSHDNMFERGYLTMNGKKIGI----------PRYYQ 245
>gi|9634955|ref|NP_054653.1| nonstructural protein [Chlamydia phage 2]
gi|7406595|emb|CAB85595.1| nonstructural protein [Chlamydia phage 2]
Length=336
Score = 87.4 bits (215), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 72/229 (31%), Positives = 114/229 (50%), Gaps = 28/229 (12%)
Query 40 FLIVPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNG-VFPEEIQL 98
++++PC +C C + A+ WS+R + E+ + Q F+TLTY + HLP+NG + + +L
Sbjct 71 WVLMPCRRCKFCRVQNAKIWSYRCMHEA-SLYSQNCFLTLTYEDRHLPENGSLVRDHPRL 129
Query 99 FFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWN--FPDNFESAYSR---LTL 153
F RLR + H +RY EYG +RPHYH++++N FPD + R L +
Sbjct 130 FLMRLREHI----YPHKIRYFGCGEYGSKLQRPHYHLLIYNYDFPDKKLLSKKRGNPLFV 185
Query 154 IESCWRRPTGEYHPDGSPVTRSIGFA--YCVPVINGGINYVMKYMGKRECAPEGMNSTFM 211
E R + GS +S G+ Y + +NG I+ + G+R PE F+
Sbjct 186 SEKLMRLWPFGFSTVGSVTRQSAGYVARYSLKKVNGDIS--QDHYGQR--LPE-----FL 236
Query 212 LASRKNGGIGSAYAEQLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYY 260
+ S K GIG+ + E+ + Q D V+ G+ T PRYY
Sbjct 237 MCSLK-PGIGADWYEKYKCDVYPQ----DYLVVQD-KGKSFKTRPPRYY 279
>gi|649562725|gb|KDS68909.1| hypothetical protein M096_3339 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=250
Score = 85.5 bits (210), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 73/232 (31%), Positives = 99/232 (43%), Gaps = 55/232 (24%)
Query 76 FITLTYNNEHLPK------------NGVFPEEIQLFFKRLRTKLDRRGISHNLRYIAVSE 123
F+TLTY++EH+P V +IQLF KRLR K + LRY SE
Sbjct 13 FVTLTYDDEHIPTAMIGEDLFKTTVGVVSKRDIQLFMKRLRKKY----AQYRLRYFLTSE 68
Query 124 YGHWSKRPHYHIILWNFPDNFESAYSRLTLIESCWRRPTGEYHPDGSPVTRSIGFAYCVP 183
YG RPHYH+IL+ FP F + L+ CW+ GF P
Sbjct 69 YGSQGGRPHYHMILFGFP--FTGKHGG-DLLAECWKN----------------GFVQAHP 109
Query 184 VINGGINYVMKYMGKRECAPEGMNST-----FMLASRKNGGIGSAYA-EQLRAFYEQQPD 237
+ I+YV KYM ++ P+ + FML S K GIG + EQ+ FY P
Sbjct 110 LTTKEISYVTKYMYEKSMIPDILKGVKEYQPFMLCS-KMPGIGYHFLREQILDFYRLHP- 167
Query 238 TCDMSVINIYTGQPLTTMLPRYYRMKFMPSTSMCYDSNFIKYFKDTVRWFEI 289
+ + G + +PRYY K YD + +Y K+ F I
Sbjct 168 ---RDYVRAFNGMRMA--MPRYYADK-------LYDDDMKEYLKELREAFFI 207
>gi|9791179|ref|NP_063900.1| conserved hypothetical protein [Chlamydia pneumoniae phage CPAR39]
gi|7190960|gb|AAF39720.1| conserved hypothetical protein [Chlamydia pneumoniae phage CPAR39]
Length=327
Score = 86.3 bits (212), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 72/229 (31%), Positives = 111/229 (48%), Gaps = 28/229 (12%)
Query 40 FLIVPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNG-VFPEEIQL 98
++++PC KC C + A+ WS+R + E+ + Q F+TLTY + +LP+NG + +L
Sbjct 62 WVVMPCLKCRFCRVRNAKIWSYRCMHEA-SLYSQNCFLTLTYEDRYLPENGSLVRNHPRL 120
Query 99 FFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWN--FPDNFESAYSR---LTL 153
F RLR ++ H +RY EYG +RPHYH++++N FPD + R L +
Sbjct 121 FLMRLRKEI----YPHKIRYFGCGEYGSKLQRPHYHLLIYNYDFPDKKLLSKKRGNPLFV 176
Query 154 IESCWRRPTGEYHPDGSPVTRSIGFA--YCVPVINGGINYVMKYMGKRECAPEGMNSTFM 211
E R + GS +S G+ Y + +NG I+ + G+R PE F+
Sbjct 177 SEKLMRLWPFGFSTVGSVTRQSAGYVARYSLKKVNGDIS--QDHYGQR--LPE-----FL 227
Query 212 LASRKNGGIGSAYAEQLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYY 260
+ S K G Y + R Y Q D V+ G+ T PRYY
Sbjct 228 MCSLKPGIGADWYEKYKRDVYPQ-----DYLVVQD-KGKSFKTRPPRYY 270
>gi|17402855|ref|NP_510879.1| hypothetical protein PhiCPG1p9 [Guinea pig Chlamydia phage]
Length=263
Score = 85.1 bits (209), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 72/226 (32%), Positives = 109/226 (48%), Gaps = 28/226 (12%)
Query 43 VPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNG-VFPEEIQLFFK 101
+P KC C + A+ WS+R + E+ + Q F+TLTY + HLP+NG + + LF
Sbjct 1 MPWRKCKFCRVQNAKIWSYRCIHEA-SLYSQNCFLTLTYEDRHLPENGSLVRDHPALFLM 59
Query 102 RLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWN--FPDNFESAYSR---LTLIES 156
RLR ++ H +RY EYG +RPHYH++++N FPD + R L + E
Sbjct 60 RLRKEI----YPHKIRYFGCGEYGSKLQRPHYHLLIYNYDFPDKKLLSKKRGNPLFVSEK 115
Query 157 CWRRPTGEYHPDGSPVTRSIGFA--YCVPVINGGINYVMKYMGKRECAPEGMNSTFMLAS 214
R + GS + +S G+ Y + +NG I+ + G+R P+ F++ S
Sbjct 116 LMRLWPFGFSTVGSVMRQSAGYVARYSLKKVNGDIS--QDHYGQR--LPQ-----FLMCS 166
Query 215 RKNGGIGSAYAEQLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYY 260
K G Y + R Y Q D V+ G+ TT PRYY
Sbjct 167 LKPGIGADWYEKYKRDVYPQ-----DYLVVQD-KGKSFTTRPPRYY 206
>gi|77020121|ref|YP_338244.1| putative replication protein [Chlamydia phage 4]
gi|59940020|gb|AAX12549.1| putative replication protein [Chlamydia phage 4]
Length=315
Score = 85.1 bits (209), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 68/227 (30%), Positives = 107/227 (47%), Gaps = 24/227 (11%)
Query 40 FLIVPCNKCALCNEKKAQQWSFRALCESYTSNKQAYFITLTYNNEHLPKNG-VFPEEIQL 98
++++PC +C C + A+ WS+R + E+ + Q F+TLTY ++HLP+NG + L
Sbjct 50 WILMPCRRCKFCRVQNAKIWSYRCMHEA-SLYSQNCFLTLTYEDQHLPENGSLVRNHPTL 108
Query 99 FFKRLRTKLDRRGISHNLRYIAVSEYGHWSKRPHYHIILWN--FPDNFESAYSR---LTL 153
F +RLR + H +RY EYG +RPHYH++++N FPD + R L +
Sbjct 109 FLRRLREHIS----PHKIRYFGCGEYGSKLQRPHYHLLIYNYDFPDKKLLSKKRGNPLFV 164
Query 154 IESCWRRPTGEYHPDGSPVTRSIGFAYCVPVINGGINYVMKYMGKRECAPEGMNSTFMLA 213
E + + GS +S G+ + + + G+R PE F++
Sbjct 165 SEKLMQLWPYGFSTVGSVTRQSAGYVARYSLKKVSRDISQDHYGQR--LPE-----FLMC 217
Query 214 SRKNGGIGSAYAEQLRAFYEQQPDTCDMSVINIYTGQPLTTMLPRYY 260
S K G Y + R Y Q D V+ G+ TT PRYY
Sbjct 218 SLKPGIGADWYEKYKRDVYPQ-----DYLVVQD-KGKSFTTRPPRYY 258
Lambda K H a alpha
0.323 0.137 0.428 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2764229385792