bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_2
Length=332
Score E
Sequences producing significant alignments: (Bits) Value
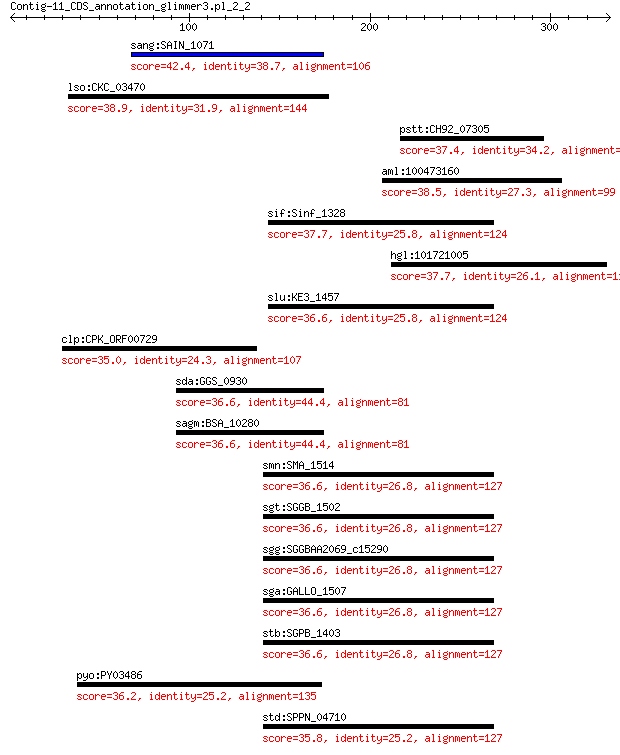
sang:SAIN_1071 hypothetical protein 42.4 0.074
lso:CKC_03470 hypothetical protein 38.9 1.5
pstt:CH92_07305 biopolymer transporter TolR 37.4 1.6
aml:100473160 myomesin-3-like 38.5 2.1
sif:Sinf_1328 HAD-superfamily hydrolase / phosphatase 37.7 2.4
hgl:101721005 Myom3; myomesin 3 37.7
slu:KE3_1457 hypothetical protein 36.6 5.3
clp:CPK_ORF00729 hypothetical protein 35.0 5.3
sda:GGS_0930 hypothetical protein 36.6 5.6
sagm:BSA_10280 hypothetical protein 36.6 5.6
smn:SMA_1514 yidA; phosphatase YidA 36.6 5.7
sgt:SGGB_1502 haloacid dehalogenase-like hydrolase 36.6 6.4
sgg:SGGBAA2069_c15290 haloacid dehalogenase-like hydrolase (EC... 36.6 6.4
sga:GALLO_1507 HAD-superfamily hydrolase / phosphatase 36.6 6.4
stb:SGPB_1403 haloacid dehalogenase-like hydrolase 36.6 6.5
pyo:PY03486 hypothetical protein 36.2 9.4
std:SPPN_04710 phosphatase YidA 35.8 9.7
> sang:SAIN_1071 hypothetical protein
Length=288
Score = 42.4 bits (98), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 41/126 (33%), Positives = 58/126 (46%), Gaps = 31/126 (25%)
Query 68 FMTLTISDENYEILKNTCKSEDKNTIATKAIRLTLERIRKKTGKSIKHWFITELGHEKTE 127
F TLT D N + D A K +R L+ R+K GK ++ F+ EL K+
Sbjct 73 FWTLTFDD-------NKVDARD-YPYAKKRLRAWLKYQREKYGK-FQYIFVAEL--HKSG 121
Query 128 RLHLHGIVWGIG----------TDQLIKEK---------WNYGITYTGNFVN-EKTINYI 167
R+H HG+ G T++LIK+K W G + + EKT NYI
Sbjct 122 RIHFHGLTAGFSPPLTEARSPKTNRLIKKKGLQIYNAETWKNGFSTVSKIQDREKTANYI 181
Query 168 TKYMTK 173
+KY+TK
Sbjct 182 SKYITK 187
> lso:CKC_03470 hypothetical protein
Length=424
Score = 38.9 bits (89), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 46/192 (24%), Positives = 73/192 (38%), Gaps = 52/192 (27%)
Query 33 YITAACGKCLECRKQKQREWLVRMSEELRTEPNAYFMTLTISDENYEILKN--------- 83
YI A C +C C K + WL R E+ +F+TLT S N+ +KN
Sbjct 222 YILARCRRCSVCCKSRGMFWLRRAQTEVMRSSRTWFITLTFSPSNH--IKNYALTIGQYV 279
Query 84 -TCKSEDKNTIATK-------------------------------AIRLTLERIRKKTGK 111
+ ED+N K I L L+R+RK T K
Sbjct 280 ESLSIEDRNFFYGKKKYGTIIEDIRSLNISDVDLKFRLLCKGFGDKIVLFLKRLRKNTSK 339
Query 112 SIKHWFITELGHEKTERLHLHGIVWGIGTDQLIK-----EKW-NYGITYTGNFVNE-KTI 164
+++ + E K+ H H ++ ++L+K E+W G ++ + T
Sbjct 340 KFRYFIVFE--KHKSGNPHAHMLIHQKSGEELLKKAEIQEEWIREGFSHVRLLREDLNTA 397
Query 165 NYITKYMTKIDE 176
Y+ KY+ K D
Sbjct 398 RYVCKYLLKEDS 409
> pstt:CH92_07305 biopolymer transporter TolR
Length=153
Score = 37.4 bits (85), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 27/80 (34%), Positives = 42/80 (53%), Gaps = 4/80 (5%)
Query 217 LRNGAKINLPIYYRNKLFTEEERELLFID-KIEKGFIYVLGTKVHRDDEEYYIQLLEEGR 275
L G KI LP L TE ER++L + K + GF + LG++++ +D+ LEE R
Sbjct 38 LVQGVKIELPKVAAEALPTENERQILTLSVKADGGFYWNLGSELNTEDQTDSAIDLEEMR 97
Query 276 KKEAILYGEHNQDWEQQKYI 295
+K A + E + Q Y+
Sbjct 98 EKVAAIIAERG---DTQVYV 114
> aml:100473160 myomesin-3-like
Length=1450
Score = 38.5 bits (88), Expect = 2.1, Method: Composition-based stats.
Identities = 27/101 (27%), Positives = 55/101 (54%), Gaps = 5/101 (5%)
Query 207 EKGKTIETYRLRNGAKINLPIYYRNKLFTEEERELLFIDKIEKGFIYVLGTKVHRDDEEY 266
E+G+ +E +L A+++L I+ ++F+ R++ F+ EKG + V+ + DD+
Sbjct 1024 EQGEWLEVEKLSPAAELHL-IFNEKEIFSSPNRKINFVR--EKGLVEVVIQNLTEDDKGS 1080
Query 267 YIQLLEEGRKKEAILYGEHNQDWEQ--QKYINRLRKQKKKQ 305
Y L++G+ K I + D+++ +K + R K+KQ
Sbjct 1081 YTAQLQDGKAKNQITLALVDDDFDKLLRKAEAKRRDWKRKQ 1121
> sif:Sinf_1328 HAD-superfamily hydrolase / phosphatase
Length=269
Score = 37.7 bits (86), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 61/128 (48%), Gaps = 16/128 (13%)
Query 144 IKEKWNYGITYTGNFVNEKTI--NYITKYMTKIDEDHPEFVGKVLCSKGIGAGYIKRADA 201
++E+ +Y IT+ G V + + N IT+ MT D EF+ + L A
Sbjct 59 LREEGDYVITFNGGLVQDTSTGENIITETMTYEDYLDIEFLSRKLDVH-------MHAIT 111
Query 202 SKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVLGTKV 259
K Y + I Y + + +N+PI+YR ++ +E +++FID+ E +L +
Sbjct 112 KKGIYTANRNIGKYTVHESSLVNMPIFYRTPEEMGDKEIIKMMFIDEPE-----ILDAAI 166
Query 260 HRDDEEYY 267
+ +E++
Sbjct 167 KKIPQEFF 174
> hgl:101721005 Myom3; myomesin 3
Length=1440
Score = 37.7 bits (86), Expect = 4.1, Method: Composition-based stats.
Identities = 31/120 (26%), Positives = 62/120 (52%), Gaps = 6/120 (5%)
Query 212 IETYRLRNGAKINLPIYYRNKLFTEEERELLFIDKIEKGFIYVLGTKVHRDDEEYYIQLL 271
+E +L A+++L I+ ++F+ R++ F D+ EKG + V+ + DD+ Y L
Sbjct 1029 LEVEKLSPAAELHL-IFNEKEIFSSPNRKINF-DR-EKGLVEVIIQHLSEDDKGSYTAQL 1085
Query 272 EEGRKKEAILYGEHNQDWEQ-QKYINRLRKQKKKQEQELNQLEIYWALERAKDYTQLETC 330
++G+ K I + D++Q Q+ + R+ K+++ Q + W + +D L TC
Sbjct 1086 QDGKAKNQITLALVDDDFDQLQRKADAKRRDWKRKQGPYFQEPLQWKV--TEDCRVLLTC 1143
> slu:KE3_1457 hypothetical protein
Length=269
Score = 36.6 bits (83), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 61/128 (48%), Gaps = 16/128 (13%)
Query 144 IKEKWNYGITYTGNFVNEKTI--NYITKYMTKIDEDHPEFVGKVLCSKGIGAGYIKRADA 201
+KE+ +Y IT+ G V + + N IT+ MT D EF+ + L A
Sbjct 59 LKEEGDYVITFNGGLVQDTSTGENIITETMTYEDYLDIEFLSRKLDVH-------MHAIT 111
Query 202 SKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVLGTKV 259
+ Y + I Y + + +N+PI+YR ++ +E ++++ID+ E VL +
Sbjct 112 KQGIYTANRNIGKYTVHESSLVNMPIFYRTPEEMGDKEIIKMMYIDEPE-----VLDAAI 166
Query 260 HRDDEEYY 267
+ +E++
Sbjct 167 EKIPQEFF 174
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 35.0 bits (79), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 47/108 (44%), Gaps = 11/108 (10%)
Query 30 RTAYITAACGKCLECRKQKQREWLVRMSEELRTEPNAYFMTLTISDENYEILKNTCKSED 89
R ++ C KC CR Q + W R E F+TLT D++
Sbjct 16 RNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHL---------PQ 66
Query 90 KNTIATKAIRLTLERIRKK-TGKSIKHWFITELGHEKTERLHLHGIVW 136
++ ++L L+R+R + + I+++ E G K +R H H +++
Sbjct 67 YGSLVKLHLQLFLKRLRDRISPHKIRYFGCGEYG-TKLQRPHYHLLIF 113
> sda:GGS_0930 hypothetical protein
Length=272
Score = 36.6 bits (83), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 36/101 (36%), Positives = 47/101 (47%), Gaps = 23/101 (23%)
Query 93 IATKAIRLTLERIRKKTGKSIKHWFITELGHEKTERLHLHGIVWGIG----------TDQ 142
I K IR L+ R+K GK ++ FI EL K RLH HG+ G T +
Sbjct 91 IVKKKIRTWLKAQREKYGK-FRYIFIPEL--HKNGRLHFHGVTGGFSPKLTKARNTKTGR 147
Query 143 LIK---------EKWNYGI-TYTGNFVNEKTINYITKYMTK 173
LIK + + G T T ++K NYITKY+TK
Sbjct 148 LIKKNGKQVYNVDSYQLGFSTVTKIDSSKKVANYITKYITK 188
> sagm:BSA_10280 hypothetical protein
Length=272
Score = 36.6 bits (83), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 36/101 (36%), Positives = 47/101 (47%), Gaps = 23/101 (23%)
Query 93 IATKAIRLTLERIRKKTGKSIKHWFITELGHEKTERLHLHGIVWGIG----------TDQ 142
I K IR L+ R+K GK ++ FI EL K RLH HG+ G T +
Sbjct 91 IVKKKIRTWLKAQREKYGK-FRYIFIPEL--HKNGRLHFHGVTGGFSPKLTKARNTKTGR 147
Query 143 LIK---------EKWNYGI-TYTGNFVNEKTINYITKYMTK 173
LIK + + G T T ++K NYITKY+TK
Sbjct 148 LIKKNGKQVYNVDSYQLGFSTVTKIDSSKKVANYITKYITK 188
> smn:SMA_1514 yidA; phosphatase YidA
Length=269
Score = 36.6 bits (83), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 34/132 (26%), Positives = 61/132 (46%), Gaps = 17/132 (13%)
Query 141 DQL-IKEKWNYGITYTGNFVNEKTI--NYITKYMTKIDEDHPEFVGKVLCSKGIGAGYIK 197
D+L +KE +Y IT+ G V + N IT+ MT D EF+ + L
Sbjct 55 DELNLKESGDYVITFNGGLVQDTATGENIITEAMTYDDYLDIEFLSRKLDVH-------M 107
Query 198 RADASKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVL 255
A + + + I Y + +N+PI+YR ++ +E +++FID+ E +L
Sbjct 108 HAITKEGIFTANRNIGKYTVHESTLVNMPIFYRTPEEMGDKEIIKMMFIDEPE-----IL 162
Query 256 GTKVHRDDEEYY 267
+ + +E+Y
Sbjct 163 DAAIAKIPQEFY 174
> sgt:SGGB_1502 haloacid dehalogenase-like hydrolase
Length=269
Score = 36.6 bits (83), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 34/132 (26%), Positives = 61/132 (46%), Gaps = 17/132 (13%)
Query 141 DQL-IKEKWNYGITYTGNFVNEKTI--NYITKYMTKIDEDHPEFVGKVLCSKGIGAGYIK 197
D+L +KE +Y IT+ G V + N IT+ MT D EF+ + L
Sbjct 55 DELNLKESGDYVITFNGGLVQDTATGENIITETMTYDDYLDIEFLSRKLDVH-------M 107
Query 198 RADASKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVL 255
A + + + I Y + +N+PI+YR ++ +E +++FID+ E +L
Sbjct 108 HAITKEGIFTANRNIGKYTVHESTLVNMPIFYRTPEEMGDKEIIKMMFIDEPE-----IL 162
Query 256 GTKVHRDDEEYY 267
+ + +E+Y
Sbjct 163 DAAIAKIPQEFY 174
> sgg:SGGBAA2069_c15290 haloacid dehalogenase-like hydrolase (EC:3.6.1.-)
Length=269
Score = 36.6 bits (83), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 34/132 (26%), Positives = 61/132 (46%), Gaps = 17/132 (13%)
Query 141 DQL-IKEKWNYGITYTGNFVNEKTI--NYITKYMTKIDEDHPEFVGKVLCSKGIGAGYIK 197
D+L +KE +Y IT+ G V + N IT+ MT D EF+ + L
Sbjct 55 DELNLKESGDYVITFNGGLVQDTATGENIITETMTYDDYLDIEFLSRKLDVH-------M 107
Query 198 RADASKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVL 255
A + + + I Y + +N+PI+YR ++ +E +++FID+ E +L
Sbjct 108 HAITKEGIFTANRNIGKYTVHESTLVNMPIFYRTPEEMGDKEIIKMMFIDEPE-----IL 162
Query 256 GTKVHRDDEEYY 267
+ + +E+Y
Sbjct 163 DAAIAKIPQEFY 174
> sga:GALLO_1507 HAD-superfamily hydrolase / phosphatase
Length=269
Score = 36.6 bits (83), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 34/132 (26%), Positives = 61/132 (46%), Gaps = 17/132 (13%)
Query 141 DQL-IKEKWNYGITYTGNFVNEKTI--NYITKYMTKIDEDHPEFVGKVLCSKGIGAGYIK 197
D+L +KE +Y IT+ G V + N IT+ MT D EF+ + L
Sbjct 55 DELNLKESGDYVITFNGGLVQDTATGENIITETMTYDDYLDIEFLSRKLDVH-------M 107
Query 198 RADASKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVL 255
A + + + I Y + +N+PI+YR ++ +E +++FID+ E +L
Sbjct 108 HAITKEGIFTANRNIGKYTVHESTLVNMPIFYRTPEEMGDKEIIKMMFIDEPE-----IL 162
Query 256 GTKVHRDDEEYY 267
+ + +E+Y
Sbjct 163 DAAIAKIPQEFY 174
> stb:SGPB_1403 haloacid dehalogenase-like hydrolase
Length=269
Score = 36.6 bits (83), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 34/132 (26%), Positives = 61/132 (46%), Gaps = 17/132 (13%)
Query 141 DQL-IKEKWNYGITYTGNFVNEKTI--NYITKYMTKIDEDHPEFVGKVLCSKGIGAGYIK 197
D+L +KE +Y IT+ G V + N IT+ MT D EF+ + L
Sbjct 55 DELNLKESGDYVITFNGGLVQDTATGENIITETMTYDDYLDIEFLSRKLDVH-------M 107
Query 198 RADASKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVL 255
A + + + I Y + +N+PI+YR ++ +E +++FID+ E +L
Sbjct 108 HAITKEGIFTANRNIGKYTVHESTLVNMPIFYRTPEEMGDKEIIKMMFIDEPE-----IL 162
Query 256 GTKVHRDDEEYY 267
+ + +E+Y
Sbjct 163 DAAIAKIPQEFY 174
> pyo:PY03486 hypothetical protein
Length=365
Score = 36.2 bits (82), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 34/150 (23%), Positives = 67/150 (45%), Gaps = 17/150 (11%)
Query 38 CGKCLECRKQKQREWL-VRMSEELRTEPNAYFMTLTISDENYEILKNTCK--SEDKNTIA 94
C KCLE K+ +++ + +S+ + P Y+ L+ ++Y KN CK + D N I
Sbjct 204 CDKCLENVKEFVKKYDELNISDITKDSP--YYQVLSTLSDDYNNFKNYCKVNNVDCNHIQ 261
Query 95 TKA-IRLTLERIRKKTGKSIKHWFITELGHEKTERLHL-----------HGIVWGIGTDQ 142
T + I++ ++ ++H+ T T +L + GI + + +
Sbjct 262 TLSPIKIKENGVQGSAHNDVQHFEATSSSSSITNKLFIVLSIFGAIAFFLGISYKVNNKE 321
Query 143 LIKEKWNYGITYTGNFVNEKTINYITKYMT 172
L + Y Y VN+K + ++T Y++
Sbjct 322 LKNITFKYYFNYIYANVNKKIVRFLTFYIS 351
> std:SPPN_04710 phosphatase YidA
Length=268
Score = 35.8 bits (81), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 32/131 (24%), Positives = 59/131 (45%), Gaps = 16/131 (12%)
Query 141 DQLIKEKWNYGITYTGNFVNEKTINY--ITKYMTKIDEDHPEFVGKVLCSKGIGAGYIKR 198
D ++++ +Y +T+ G V E + I++ +T D EF+ + L G
Sbjct 56 DLQLRDEGDYVVTFNGALVQETATGHEIISESLTYEDYLDMEFLSRKL-------GVHMH 108
Query 199 ADASKHKYEKGKTIETYRLRNGAKINLPIYYRN--KLFTEEERELLFIDKIEKGFIYVLG 256
A Y + I TY + +N+PI+YR ++ +E + +FID+ E VL
Sbjct 109 AITKDGIYTANRNIGTYTVHESTLVNMPIFYRTPEEMADKEIVKCMFIDEPE-----VLD 163
Query 257 TKVHRDDEEYY 267
+ + E+Y
Sbjct 164 AAIEKIPAEFY 174
Lambda K H a alpha
0.317 0.136 0.408 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 600634975296