bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_6
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
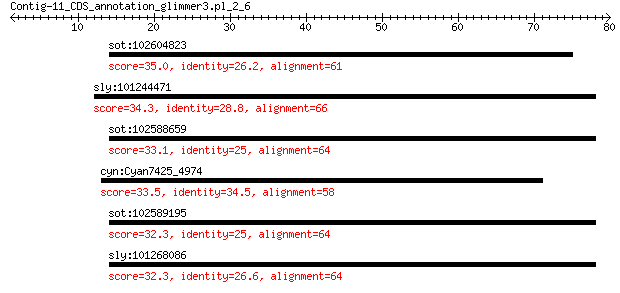
sot:102604823 1-aminocyclopropane-1-carboxylate oxidase 5-like 35.0 0.93
sly:101244471 1-aminocyclopropane-1-carboxylate oxidase 1-like 34.3 1.5
sot:102588659 1-aminocyclopropane-1-carboxylate oxidase 5-like 33.1 3.9
cyn:Cyan7425_4974 PAS/PAC and GAF sensor-containing diguanylat... 33.5 4.0
sot:102589195 1-aminocyclopropane-1-carboxylate oxidase 5-like 32.3 7.3
sly:101268086 flavonol synthase/flavanone 3-hydroxylase-like 32.3 7.8
> sot:102604823 1-aminocyclopropane-1-carboxylate oxidase 5-like
Length=308
Score = 35.0 bits (79), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 27/61 (44%), Gaps = 0/61 (0%)
Query 14 PESEEFIITIGNHLATAEKFKSRKAAEMRINKTDWNLVSALFFALKEADKWAEKNPELVK 73
P ++ ++ IG+ + + + A I D N S FF + DKW E P+ K
Sbjct 205 PSKDKLVVNIGDVIQVLSNNRFKSATHRVIRPRDANRFSCTFFYNVQGDKWVEPLPQFTK 264
Query 74 Q 74
+
Sbjct 265 E 265
> sly:101244471 1-aminocyclopropane-1-carboxylate oxidase 1-like
Length=308
Score = 34.3 bits (77), Expect = 1.5, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 32/67 (48%), Gaps = 1/67 (1%)
Query 12 PLPESEE-FIITIGNHLATAEKFKSRKAAEMRINKTDWNLVSALFFALKEADKWAEKNPE 70
P+P S++ ++ IG+ + K + A + + N S +FF E DKW E P+
Sbjct 202 PIPPSKDKLVVNIGDVIQVLSNNKFKSATHKVVRPIETNRYSYVFFYNVEGDKWIEPLPQ 261
Query 71 LVKQNKE 77
K+ E
Sbjct 262 FTKEIGE 268
> sot:102588659 1-aminocyclopropane-1-carboxylate oxidase 5-like
Length=309
Score = 33.1 bits (74), Expect = 3.9, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 29/64 (45%), Gaps = 0/64 (0%)
Query 14 PESEEFIITIGNHLATAEKFKSRKAAEMRINKTDWNLVSALFFALKEADKWAEKNPELVK 73
P ++ ++ IG+ + K + A + ++ N S +FF DKW E P+ K
Sbjct 206 PSKDKLVVNIGDVIQVLSNNKFKSATHKVVRSSETNRYSYVFFYNVHGDKWVEPLPQFTK 265
Query 74 QNKE 77
+ E
Sbjct 266 EIGE 269
> cyn:Cyan7425_4974 PAS/PAC and GAF sensor-containing diguanylate
cyclase/phosphodiesterase
Length=877
Score = 33.5 bits (75), Expect = 4.0, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 33/64 (52%), Gaps = 6/64 (9%)
Query 13 LPESEEFIITIGNHLATAEKFKSRKAAEMRINKTDWNLVSAL------FFALKEADKWAE 66
LPE + F I + +A A + + R+ AE + KT+ L + L FF+L E ++
Sbjct 159 LPEEQSFAGAIADMVALALEARDRQQAEAELQKTNQQLRNILESLTSGFFSLNETAEFTY 218
Query 67 KNPE 70
NP+
Sbjct 219 INPQ 222
> sot:102589195 1-aminocyclopropane-1-carboxylate oxidase 5-like
Length=308
Score = 32.3 bits (72), Expect = 7.3, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 28/64 (44%), Gaps = 0/64 (0%)
Query 14 PESEEFIITIGNHLATAEKFKSRKAAEMRINKTDWNLVSALFFALKEADKWAEKNPELVK 73
P + ++ IG+ + K + A + ++ N S +FF DKW E P+ K
Sbjct 205 PSKNKLVVNIGDVIQVLSNNKFKSATHKVVRPSETNRYSYVFFYNVHGDKWVEPLPQFTK 264
Query 74 QNKE 77
+ E
Sbjct 265 EIGE 268
> sly:101268086 flavonol synthase/flavanone 3-hydroxylase-like
Length=309
Score = 32.3 bits (72), Expect = 7.8, Method: Composition-based stats.
Identities = 17/64 (27%), Positives = 28/64 (44%), Gaps = 0/64 (0%)
Query 14 PESEEFIITIGNHLATAEKFKSRKAAEMRINKTDWNLVSALFFALKEADKWAEKNPELVK 73
P ++ ++ IG+ + K + A I + N S FF + DKW E P+ K
Sbjct 206 PSKDKLVVNIGDVIQVLSNNKFKSATHRVIRPRETNRYSCAFFYNVQGDKWVEPLPQCTK 265
Query 74 QNKE 77
+ E
Sbjct 266 EIGE 269
Lambda K H a alpha
0.313 0.128 0.359 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127888521377