bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-11_CDS_annotation_glimmer3.pl_2_7
Length=648
Score E
Sequences producing significant alignments: (Bits) Value
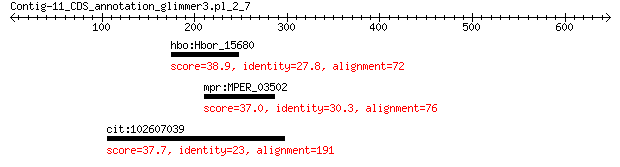
hbo:Hbor_15680 glycine/d-amino acid oxidase, deaminating 38.9 3.2
mpr:MPER_03502 hypothetical protein 37.0 6.3
cit:102607039 70 kDa peptidyl-prolyl isomerase-like 37.7 8.4
> hbo:Hbor_15680 glycine/d-amino acid oxidase, deaminating
Length=376
Score = 38.9 bits (89), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 20/72 (28%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 175 FDIFKNYYANTQEENFYIIGATETITNIKVTQAEGAIYTSTTPDKINIGIASGDSVEITP 234
+D+ +++ A Q + +++ I + + QA G + P + +A G TP
Sbjct 130 YDLPEDHRAVYQPDGGFLVPEQCIIAHTEAAQAAGGEIRAREPMRDFTPLADGGVRVTTP 189
Query 235 KNTYEADELIIT 246
K TYEAD L++T
Sbjct 190 KGTYEADRLVVT 201
> mpr:MPER_03502 hypothetical protein
Length=163
Score = 37.0 bits (84), Expect = 6.3, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 40/79 (51%), Gaps = 5/79 (6%)
Query 210 AIYTSTTPDKINIGIASGDSVEITPKNTYEAD--ELIITWFDMSTETTRTGKPTEFGTWS 267
A+ T + P + +++ P +T EA + +ITW+D + RT + G W+
Sbjct 36 ALPTESRPSAVTWWTKRARTIKGVPPSTTEAQFPQSVITWWDSMMPSWRT--QNQDGHWA 93
Query 268 KT-SGNWNVVMAPTQVGLL 285
+T G+W ++ P Q GLL
Sbjct 94 QTGEGDWGILHCPGQNGLL 112
> cit:102607039 70 kDa peptidyl-prolyl isomerase-like
Length=606
Score = 37.7 bits (86), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 44/204 (22%), Positives = 84/204 (41%), Gaps = 19/204 (9%)
Query 106 GLNMSDIKLPQLSISINKTKDNPTPDNQWSQINPSCLLAYLGIRGYAYNTTTTSQEVTVQ 165
G N+SDI + P + ++ S + L G ++T EVT+
Sbjct 12 GANLSDIDGEEEEPGEVIESAPPLKVGEERELGNSGIKKKLLKNGVGWDTPEFGDEVTIH 71
Query 166 KMAVPLLG-YFDIFKNYYANTQEENFYIIGATETITN-----IKVTQAEGAIYTSTTPDK 219
+ L G FD ++ Y + +G + T I + + E A++T T P +
Sbjct 72 YVGTLLDGTKFDSTRDRYDPLT----FKLGTGQVATGLDNGIITMKKRECAVFTFTLPSE 127
Query 220 INIGIASGDSVEITPKNTYEADELIITWF---DMSTETTRTGKPTEFGTWSKTSGNWNVV 276
+ G+ DS + P + + + +++W D+S + K E G + G+ + V
Sbjct 128 LRFGVEGRDS--LPPNSVVQFEVELVSWITVVDLSKDGGIVKKILEKGERDASPGDLDEV 185
Query 277 MAPTQV----GLLMSITPKNRVQL 296
+ QV G +++ TP+ V+
Sbjct 186 LVKYQVMLGDGTMVAKTPEEGVEF 209
Lambda K H a alpha
0.315 0.132 0.393 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1527517915246