bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_5
Length=589
Score E
Sequences producing significant alignments: (Bits) Value
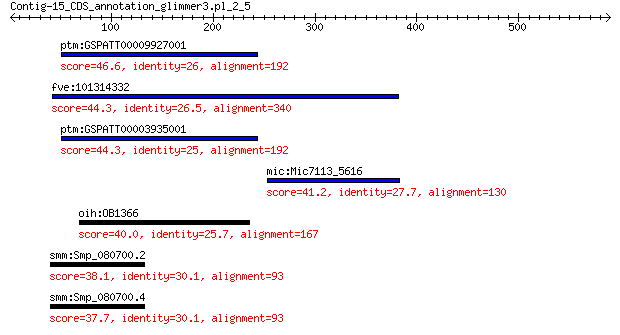
ptm:GSPATT00009927001 hypothetical protein 46.6 0.013
fve:101314332 capsid protein VP1-like 44.3 0.056
ptm:GSPATT00003935001 hypothetical protein 44.3 0.068
mic:Mic7113_5616 hypothetical protein 41.2 0.85
oih:OB1366 hypothetical protein 40.0 1.1
smm:Smp_080700.2 jnk/sapk-associated protein 38.1 7.6
smm:Smp_080700.4 jnk/sapk-associated protein 37.7 7.9
> ptm:GSPATT00009927001 hypothetical protein
Length=519
Score = 46.6 bits (109), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 50/205 (24%), Positives = 82/205 (40%), Gaps = 26/205 (13%)
Query 51 DLKSFTRTQPLNTAAFARM--REYYDFYFVPYDLLWN---KANTSLTQMYDNPQHALDSS 105
DL+ +N A R+ RE + + D+L+ K L Q+ D Q LDS
Sbjct 173 DLQQLNSLAEMNDATLKRVITREEQEILQI-QDVLYQRISKLQQKLIQLNDIEQKQLDSK 231
Query 106 PLNVVKLDGSMPFTDLSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGY--G 163
N K+D DL + N L NK + A L++CLGY
Sbjct 232 MNNNNKVDQIQDREDL-----FENQLDEPKQQEQNKGSS-----QQIRAMLIQCLGYQVS 281
Query 164 NLYYYAESTSNTFAKRPLHYNLNVSLFNL------LAYQKIYADYYRDSQWERVSPSCFN 217
++YY + TF + N+ + N L Q I+ ++YR+ Q R+ S
Sbjct 282 SIYYDKKRYQITFTYK--QANVQYTFQNQKQIDGNLEIQDIFNEFYRELQENRLVLSGLK 339
Query 218 VDYMPYSTGSSGMMLSFNYSDFYEN 242
+DY + + G ++ + ++N
Sbjct 340 IDYKQKTEINKGEIVQLSLKHQFQN 364
> fve:101314332 capsid protein VP1-like
Length=421
Score = 44.3 bits (103), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 90/361 (25%), Positives = 139/361 (39%), Gaps = 92/361 (25%)
Query 42 VLPGDTFKIDLKSFTRTQPLNTAAFARMREYYD---FYFVPYDLLWNKANTSLTQMYDNP 98
VLPGDTF +++ F R L T F M + F+FVP L+WN + + DNP
Sbjct 77 VLPGDTFNVNVTMFGR---LATPIFPVMDNLHLDSFFFFVPNRLVWNNWVKFMGEQ-DNP 132
Query 99 QHALDSSPLNVVKLDGSMPFTDLSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLME 158
++ S V G AV + +YFG L +A +
Sbjct 133 ADSISYSIPQQVSPAGGY--------------------AVGSLQDYFG----LPTAGQV- 167
Query 159 CLGYGNLYYYAESTSNTFAKRPLHYNLNVSLFNLLAYQKIYADYYRDSQWERVSPSCFNV 218
SNT + H L V +NL IY ++RD + + +
Sbjct 168 ------------GVSNTVS----HSALPVRAYNL-----IYNQWFRDENLQ--NSVVVDK 204
Query 219 DYMPYSTGSSGMMLSFNYSDFYENYSMFDLRYCNWQKDLFHGVVPNQQYGDVA---SISM 275
P +T S+ NY++ LR + D F +P Q G A +
Sbjct 205 GDGPDTTPST-------------NYTL--LRRGK-RHDYFTSALPWPQKGGTAVSLPLGT 248
Query 276 SVPVV----AGSSAALINS-------RITSSNSTTTLRFPTDPAIPDATPLLTHPSFSIL 324
S P+ +GS +I++ + S+ S T+L+ + AT L S +
Sbjct 249 SAPIAFSGASGSDVGVISTTQGNLIKNMYSTGSGTSLKIGSATV---ATGLYADLSAATA 305
Query 325 A----LRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCTYLGGISSSLDINE 380
A LRQ+ +QK E G Y E + H+ V+ D + YLGG S+ ++I
Sbjct 306 ATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINIAP 365
Query 381 V 381
+
Sbjct 366 I 366
> ptm:GSPATT00003935001 hypothetical protein
Length=519
Score = 44.3 bits (103), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 48/205 (23%), Positives = 83/205 (40%), Gaps = 27/205 (13%)
Query 51 DLKSFTRTQPLNTAAFARM--REYYDFYFVPYDLLWNKA---NTSLTQMYDNPQHALDSS 105
+L+ + +N A R+ RE + + DLL K L Q+ + Q LDS
Sbjct 174 ELQQLSSLAEMNEATLKRVITREEQEILII-QDLLTQKISKLQQKLIQLNEIEQKQLDSR 232
Query 106 PLNVVKLDGSMPFTDLSSISRYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGY--G 163
N KL+ ++ N K G ++ + A L++CLGY
Sbjct 233 MSNNNKLNQLQERENI----------IENQLDEPKKQEQKGSSQQI-RAILIQCLGYKVS 281
Query 164 NLYYYAESTSNTFAKRPLHYNLNVSLFNL------LAYQKIYADYYRDSQWERVSPSCFN 217
++YY + TF + N+ + N L Q+I+ ++YR+ Q R+ S
Sbjct 282 HIYYDKKQYQITFTYK--QANVQYTFQNQKQIDANLEVQEIFNEFYRELQENRLVLSGLK 339
Query 218 VDYMPYSTGSSGMMLSFNYSDFYEN 242
VDY + + G +L + ++N
Sbjct 340 VDYKQKTEINKGELLQLSLKHQFQN 364
> mic:Mic7113_5616 hypothetical protein
Length=1041
Score = 41.2 bits (95), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 36/135 (27%), Positives = 61/135 (45%), Gaps = 13/135 (10%)
Query 253 WQKDLFHGVVPNQQYGDVASI--SMSVPVVAGSSAALINSRITSSNS---TTTLRFPTDP 307
W + + VP+ +Y D+ + S + + S A I + I N T LR+ P
Sbjct 625 WLRYGWSDTVPSPKYQDIQKLLNSHTAAIYWHVSPAAITTFILRQNKSLKTFVLRYDQPP 684
Query 308 AIPDATPLLTHPSFSILALRQAEFLQKWKEITQSGNKDYKEQVEKHWNVSPGDGFSEMCT 367
+ A +L + +++ A R+ + L+ W + KDYK Q EK W E+ T
Sbjct 685 HLLCAETMLDNSTYAAAA-RKLQDLENWLTHWKQDYKDYKVQGEKRWR-------QEIPT 736
Query 368 YLGGISSSLDINEVV 382
L +S L+I E++
Sbjct 737 RLNELSQILNIAEIL 751
> oih:OB1366 hypothetical protein
Length=298
Score = 40.0 bits (92), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 43/183 (23%), Positives = 80/183 (44%), Gaps = 24/183 (13%)
Query 69 MREYYDFYFVPYDLLWNK-ANTSLTQMY--DNPQHALDSSPLNVVKLDGSMPFTDLSSIS 125
M+ YY F+ + LL+ K ++ ++ +Y +N + LD LN +L G+ F DL ++
Sbjct 1 MKVYYFIIFICFILLFPKVSDAAIIDLYIDENVRGNLD---LNQYQLLGNEGFFDLQKLN 57
Query 126 RYLNSLASNSTAVTNKANYFGYNRALCSAKLMECLGYG-----------NLYYYAESTSN 174
+++ L A Y + + ME +GY YY +
Sbjct 58 TFMDELQKEVNKEPKDAYYDSTGKLI-----MEKVGYTLDREKFIELIYQSYYVENNDKV 112
Query 175 TFAKRPLHYNLNVSLFNLLAYQKI--YADYYRDSQWERVSPSCFNVDYMPYSTGSSGMML 232
T KRP+H +N +L ++ ++I Y +R+S +R + + + + G +
Sbjct 113 TVPKRPVHARVNHALLREISQKQIGFYTTTFRESNTQRANNIKLAAESINNTVLFPGEVF 172
Query 233 SFN 235
SFN
Sbjct 173 SFN 175
> smm:Smp_080700.2 jnk/sapk-associated protein
Length=1210
Score = 38.1 bits (87), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 45/103 (44%), Gaps = 10/103 (10%)
Query 40 WEVLPGDTFKIDLKS---------FTRTQPLNTAAFARMREYYDFYFVPYDLLWNKANTS 90
WE+LP DTF D+++ TR + N A + E +F F+ YD N
Sbjct 671 WEMLPMDTFSGDIETGQSLTADMLETRIKKCNVNAPCSLNECTEFKFLEYDNTNNNTPKK 730
Query 91 LTQMYDNPQHALDSSPLNVVKLDGSMPFTD-LSSISRYLNSLA 132
+ ++ ++ S+P + L D LSS R +N+L
Sbjct 731 INEIVQESKYHRTSTPTSSSHLSSIKQAIDHLSSTRRLVNALG 773
> smm:Smp_080700.4 jnk/sapk-associated protein
Length=1212
Score = 37.7 bits (86), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 45/103 (44%), Gaps = 10/103 (10%)
Query 40 WEVLPGDTFKIDLKS---------FTRTQPLNTAAFARMREYYDFYFVPYDLLWNKANTS 90
WE+LP DTF D+++ TR + N A + E +F F+ YD N
Sbjct 673 WEMLPMDTFSGDIETGQSLTADMLETRIKKCNVNAPCSLNECTEFKFLEYDNTNNNTPKK 732
Query 91 LTQMYDNPQHALDSSPLNVVKLDGSMPFTD-LSSISRYLNSLA 132
+ ++ ++ S+P + L D LSS R +N+L
Sbjct 733 INEIVQESKYHRTSTPTSSSHLSSIKQAIDHLSSTRRLVNALG 775
Lambda K H a alpha
0.317 0.132 0.397 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1347988917239