bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_2
Length=298
Score E
Sequences producing significant alignments: (Bits) Value
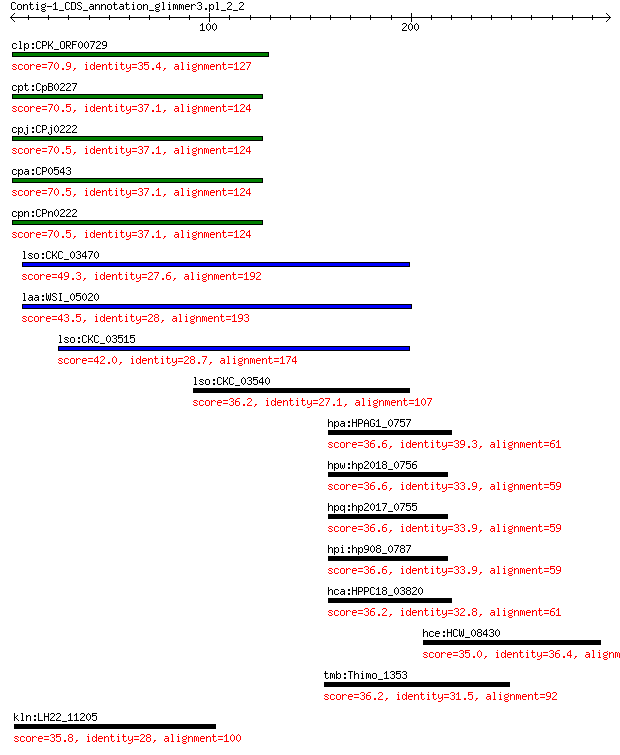
clp:CPK_ORF00729 hypothetical protein 70.9 2e-12
cpt:CpB0227 hypothetical protein 70.5 2e-12
cpj:CPj0222 hypothetical protein 70.5 2e-12
cpa:CP0543 hypothetical protein 70.5 2e-12
cpn:CPn0222 hypothetical protein 70.5 2e-12
lso:CKC_03470 hypothetical protein 49.3 5e-04
laa:WSI_05020 hypothetical protein 43.5 0.035
lso:CKC_03515 hypothetical protein 42.0 0.10
lso:CKC_03540 hypothetical protein 36.2 1.8
hpa:HPAG1_0757 N-acetylmuramoyl-L-alanine amidase (EC:3.5.1.28) 36.6 6.3
hpw:hp2018_0756 amiA; N-acetylmuramoyl-L-alanine amidase (EC:3... 36.6 6.5
hpq:hp2017_0755 amiA; N-acetylmuramoyl-L-alanine amidase (EC:3... 36.6 6.5
hpi:hp908_0787 amiA; N-acetyl muramoyl-L-alanine amidase (EC:3... 36.6 6.5
hca:HPPC18_03820 N-acetylmuramoyl-L-alanine amidase 36.2 7.4
hce:HCW_08430 F0F1 ATP synthase subunit B' (EC:3.6.3.14) 35.0 7.4
tmb:Thimo_1353 c-type cytochrome biogenesis protein CcmF 36.2 8.4
kln:LH22_11205 lipid A biosynthesis (KDO)2-(lauroyl)-lipid IVA... 35.8 8.9
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 45/130 (35%), Positives = 65/130 (50%), Gaps = 32/130 (25%)
Query 2 YQKNIMLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLTYDDEHVP--GMIVK 59
Y+ +L+PC +C CR + + W+ R EA Y K F+TLTYDD+H+P G +VK
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKN--CFLTLTYDDKHLPQYGSLVK 72
Query 60 TGEIMRKVQYVWKPGEKRPESVQTLLYTDVQKFLKRLR-KAYRGKLRYFVAGEYGEQTAR 118
+Q FLKRLR + K+RYF GEYG + R
Sbjct 73 L---------------------------HLQLFLKRLRDRISPHKIRYFGCGEYGTKLQR 105
Query 119 PHYHMILYGW 128
PHYH++++ +
Sbjct 106 PHYHLLIFNY 115
> cpt:CpB0227 hypothetical protein
Length=113
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 63/127 (50%), Gaps = 32/127 (25%)
Query 2 YQKNIMLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLTYDDEHVP--GMIVK 59
Y+ +L+PC +C CR + + W+ R EA Y E+ F+TLTYDD+H+P G +VK
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLY--EKNCFLTLTYDDKHLPQYGSLVK 72
Query 60 TGEIMRKVQYVWKPGEKRPESVQTLLYTDVQKFLKRLRKAYR-GKLRYFVAGEYGEQTAR 118
+Q FLKRLRK K+RYF G YG + R
Sbjct 73 L---------------------------HLQLFLKRLRKMISPHKIRYFECGAYGTKLQR 105
Query 119 PHYHMIL 125
PHYH++L
Sbjct 106 PHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 63/127 (50%), Gaps = 32/127 (25%)
Query 2 YQKNIMLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLTYDDEHVP--GMIVK 59
Y+ +L+PC +C CR + + W+ R EA Y E+ F+TLTYDD+H+P G +VK
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLY--EKNCFLTLTYDDKHLPQYGSLVK 72
Query 60 TGEIMRKVQYVWKPGEKRPESVQTLLYTDVQKFLKRLRKAYR-GKLRYFVAGEYGEQTAR 118
+Q FLKRLRK K+RYF G YG + R
Sbjct 73 L---------------------------HLQLFLKRLRKMISPHKIRYFECGAYGTKLQR 105
Query 119 PHYHMIL 125
PHYH++L
Sbjct 106 PHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 63/127 (50%), Gaps = 32/127 (25%)
Query 2 YQKNIMLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLTYDDEHVP--GMIVK 59
Y+ +L+PC +C CR + + W+ R EA Y E+ F+TLTYDD+H+P G +VK
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLY--EKNCFLTLTYDDKHLPQYGSLVK 72
Query 60 TGEIMRKVQYVWKPGEKRPESVQTLLYTDVQKFLKRLRKAYR-GKLRYFVAGEYGEQTAR 118
+Q FLKRLRK K+RYF G YG + R
Sbjct 73 L---------------------------HLQLFLKRLRKMISPHKIRYFECGAYGTKLQR 105
Query 119 PHYHMIL 125
PHYH++L
Sbjct 106 PHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 63/127 (50%), Gaps = 32/127 (25%)
Query 2 YQKNIMLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLTYDDEHVP--GMIVK 59
Y+ +L+PC +C CR + + W+ R EA Y E+ F+TLTYDD+H+P G +VK
Sbjct 15 YRNRWVLMPCLKCRFCRTQHAKVWSYRCVHEASLY--EKNCFLTLTYDDKHLPQYGSLVK 72
Query 60 TGEIMRKVQYVWKPGEKRPESVQTLLYTDVQKFLKRLRKAYR-GKLRYFVAGEYGEQTAR 118
+Q FLKRLRK K+RYF G YG + R
Sbjct 73 L---------------------------HLQLFLKRLRKMISPHKIRYFECGAYGTKLQR 105
Query 119 PHYHMIL 125
PHYH++L
Sbjct 106 PHYHLLL 112
> lso:CKC_03470 hypothetical protein
Length=424
Score = 49.3 bits (116), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 53/215 (25%), Positives = 86/215 (40%), Gaps = 36/215 (17%)
Query 7 MLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLTYD-DEHVPGMIVKTGEIM- 64
+L C +C C + W R + E WFITLT+ H+ + G+ +
Sbjct 223 ILARCRRCSVCCKSRGMFWLRRAQTEVM--RSSRTWFITLTFSPSNHIKNYALTIGQYVE 280
Query 65 ------RKVQYVWKPGEKRPESVQTLLYTDVQK---------------FLKRLRKAYRGK 103
R Y K E +++L +DV FLKRLRK K
Sbjct 281 SLSIEDRNFFYGKKKYGTIIEDIRSLNISDVDLKFRLLCKGFGDKIVLFLKRLRKNTSKK 340
Query 104 LRYFVAGEYGEQTARPHYHMILYGWQPTDLEHLYKIQHNGYFTSKWLADLWGMGQIQIAQ 163
RYF+ E ++ PH HM+++ +L +IQ +W+ + G +++ +
Sbjct 341 FRYFIVFE-KHKSGNPHAHMLIHQKSGEELLKKAEIQE------EWIRE--GFSHVRLLR 391
Query 164 AVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQK 198
T RYV Y+ K+ + G + A + G K
Sbjct 392 EDLNTARYVCKYLLKE--DSKGIRVRASFCYGSMK 424
> laa:WSI_05020 hypothetical protein
Length=405
Score = 43.5 bits (101), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 54/218 (25%), Positives = 87/218 (40%), Gaps = 42/218 (19%)
Query 7 MLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLT----------------YDD 50
+++PC C C + W R +E + WF+TLT Y D
Sbjct 202 LILPCRSCSSCYKNRGLFWLRRAYIEVK--RSTRTWFVTLTMTPANHFANHRSMVFNYID 259
Query 51 EHVP---GMIVKTG-----EIMRKVQYVWKPGEKRPESVQTLLYTD-VQKFLKRLRKAYR 101
P ++ G +MRK GE S+ + + V FLKRLRK
Sbjct 260 SFPPHERDLLNVDGRPTEIHLMRKKDIF---GENVLFSLLCKGFGNKVSLFLKRLRKNTG 316
Query 102 GKLRYFVAGEYGEQTARPHYHMILYGWQPTDLEHLYKIQHNGYFTSKWLADLWGMGQIQI 161
K RYF E ++ PH HM+++ Q +L ++Q +W + G +++
Sbjct 317 KKFRYFFVFE-KHKSGDPHVHMLIHQ-QCDNLLKKAEVQ------EEWSRE--GFSHVRL 366
Query 162 AQAVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQKP 199
+ T RYV Y+ K+ + G + A ++ G P
Sbjct 367 LKEDLFTARYVCKYLMKE--GMKGIRVRASFQYGALTP 402
> lso:CKC_03515 hypothetical protein
Length=432
Score = 42.0 bits (97), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 50/197 (25%), Positives = 79/197 (40%), Gaps = 36/197 (18%)
Query 25 WTTRIELEARDYPKEEVWFITLTYD-DEHVPGMIVKTGEIM-------RKVQYVWKPGEK 76
W R + E WF+TLT+ H+ + G+ + R + Y K
Sbjct 244 WLRRAQTEVM--RSSRTWFVTLTFSPSNHIKNYALVVGQYVDSLSTEDRDLFYGKKKYGT 301
Query 77 RPESVQTLLYTDVQK---------------FLKRLRKAYRGKLRYFVAGEYGEQTARPHY 121
E + L TDV FLKRLRK K RYFV E ++ PH
Sbjct 302 IFEDLTILNITDVDLKFRLLCKGFGDKIVLFLKRLRKNTSKKFRYFVVFE-RHKSGDPHA 360
Query 122 HMILYGWQPTDLEHLYKIQHNGYFTSKWLADLWGMGQIQIAQAVPETYRYVAGYVTKKMY 181
HM+++ +L +IQ +W+ + G +++ + +T RYV Y+ K+
Sbjct 361 HMLIHQKPGDELIKKAEIQ------EEWMRE--GFSHVRLLREDLKTARYVCKYLLKE-- 410
Query 182 EIDGQKANAYYELGQQK 198
+ G + A + G K
Sbjct 411 DSKGIRVRASFRYGSLK 427
> lso:CKC_03540 hypothetical protein
Length=107
Score = 36.2 bits (82), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 49/107 (46%), Gaps = 11/107 (10%)
Query 92 FLKRLRKAYRGKLRYFVAGEYGEQTARPHYHMILYGWQPTDLEHLYKIQHNGYFTSKWLA 151
FLKRLRK K RYF E ++ H HM+++ +L ++Q +W+
Sbjct 12 FLKRLRKNTSKKFRYFFVFE-KHKSGNLHAHMLIHQEIGDELLKKAEVQ------EEWMR 64
Query 152 DLWGMGQIQIAQAVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQK 198
+ G +++ + T RYV Y+ K+ + G + A + G K
Sbjct 65 E--GFSHVRLLKEDLNTARYVCKYLLKE--DAKGIRVRASFRYGSMK 107
> hpa:HPAG1_0757 N-acetylmuramoyl-L-alanine amidase (EC:3.5.1.28)
Length=432
Score = 36.6 bits (83), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 24/63 (38%), Positives = 32/63 (51%), Gaps = 4/63 (6%)
Query 159 IQIAQAVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQKPFA--CMSLKPGLGDHYYQE 216
I IAQ P+ R V GY K YE+ K Y + ++KP A M LKP H+ +
Sbjct 79 ITIAQFSPKLVRVVIGYAPKMTYEVKILKDKLYVSIVEKKPLARHQMVLKP--PKHHALK 136
Query 217 HKA 219
H+A
Sbjct 137 HQA 139
> hpw:hp2018_0756 amiA; N-acetylmuramoyl-L-alanine amidase (EC:3.5.1.28)
Length=435
Score = 36.6 bits (83), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 20/59 (34%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 159 IQIAQAVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQKPFACMSLKPGLGDHYYQEH 217
I IAQ P+ R V GY K YE+ K Y + ++KP A + P H+ +H
Sbjct 79 ITIAQFSPKLVRVVIGYAPKMTYEVKILKDKLYISIVEKKPLARHQITPKPPKHHALKH 137
> hpq:hp2017_0755 amiA; N-acetylmuramoyl-L-alanine amidase (EC:3.5.1.28)
Length=435
Score = 36.6 bits (83), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 20/59 (34%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 159 IQIAQAVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQKPFACMSLKPGLGDHYYQEH 217
I IAQ P+ R V GY K YE+ K Y + ++KP A + P H+ +H
Sbjct 79 ITIAQFSPKLVRVVIGYAPKMTYEVKILKDKLYISIVEKKPLARHQITPKPPKHHALKH 137
> hpi:hp908_0787 amiA; N-acetyl muramoyl-L-alanine amidase (EC:3.5.1.28)
Length=435
Score = 36.6 bits (83), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 20/59 (34%), Positives = 28/59 (47%), Gaps = 0/59 (0%)
Query 159 IQIAQAVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQKPFACMSLKPGLGDHYYQEH 217
I IAQ P+ R V GY K YE+ K Y + ++KP A + P H+ +H
Sbjct 79 ITIAQFSPKLVRVVIGYAPKMTYEVKILKDKLYISIVEKKPLARHQITPKPPKHHALKH 137
> hca:HPPC18_03820 N-acetylmuramoyl-L-alanine amidase
Length=431
Score = 36.2 bits (82), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 20/61 (33%), Positives = 29/61 (48%), Gaps = 0/61 (0%)
Query 159 IQIAQAVPETYRYVAGYVTKKMYEIDGQKANAYYELGQQKPFACMSLKPGLGDHYYQEHK 218
I IAQ P+ R V GY K YE+ K Y + ++KP A + P H+ +H+
Sbjct 79 ITIAQFSPKLARVVIGYAPKMTYEVKILKDKLYISIVEKKPLARHQITPKPPKHHALKHQ 138
Query 219 A 219
Sbjct 139 V 139
> hce:HCW_08430 F0F1 ATP synthase subunit B' (EC:3.6.3.14)
Length=140
Score = 35.0 bits (79), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 32/97 (33%), Positives = 50/97 (52%), Gaps = 11/97 (11%)
Query 206 KPGLGDHYYQEHKAEIWRQ-GYIQCTNGKHAQIPRYYEKMMEAENPQRLWRIKQ-NRQA- 262
KP L + +AEI I+ N + QI E +++ N QR I Q +QA
Sbjct 30 KPLLA--FMDNREAEINDSLSKIESDNKQSTQIQSQIEDLLKDANEQRRAIIAQATKQAI 87
Query 263 ----AAIA--ENRLKYENTDFAEQCKTKERVIKKQMK 293
A IA E+ L+ E DF+ Q K++++V+K+Q+K
Sbjct 88 EAYDAIIAQKESELEQEFEDFSSQLKSEKQVLKEQLK 124
> tmb:Thimo_1353 c-type cytochrome biogenesis protein CcmF
Length=661
Score = 36.2 bits (82), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 37/93 (40%), Gaps = 6/93 (6%)
Query 157 GQIQIAQAVPETYRYVAGYVTKKMYEID-GQKANAYYELGQQKPFACMSLKPGLGDHYYQ 215
G Q+A VPE YVA T ID G + Y LG+ SL+ YY+
Sbjct 563 GDRQVALLVPEKRTYVASTQTMTEASIDAGLLRDIYVSLGEPLEVGAWSLR-----IYYK 617
Query 216 EHKAEIWRQGYIQCTNGKHAQIPRYYEKMMEAE 248
+ IW G + T G A R Y +A
Sbjct 618 PYVRWIWLGGLLMATGGVLAITDRRYRAATQAS 650
> kln:LH22_11205 lipid A biosynthesis (KDO)2-(lauroyl)-lipid IVA
acyltransferase
Length=325
Score = 35.8 bits (81), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 28/110 (25%), Positives = 46/110 (42%), Gaps = 10/110 (9%)
Query 3 QKNIMLIPCGQCIGCRIRQREDWTTRIELEARDYPKEEVWFITLTYDDEHVPGMIVKT-G 61
Q +M+ G R+RQR DW R +EA KE V F+ +P M++ + G
Sbjct 96 QAMVMMAELGMRDPARVRQRVDWHGREIIEALQANKENVIFLVPHGWAVDIPAMLLASEG 155
Query 62 EIMRK---------VQYVWKPGEKRPESVQTLLYTDVQKFLKRLRKAYRG 102
++M + YVW +R ++ F+ +R+ Y G
Sbjct 156 QMMAAMFHNQSDPLLDYVWNTVRRRFGGRMHARNDGIKPFIASVRQGYYG 205
Lambda K H a alpha
0.321 0.136 0.431 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 505086094880