bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_1
Length=681
Score E
Sequences producing significant alignments: (Bits) Value
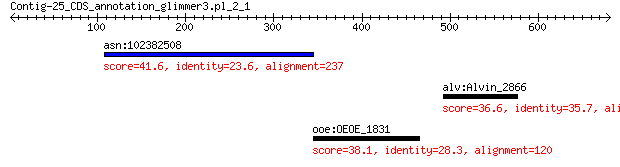
asn:102382508 endonuclease domain-containing 1 protein-like 41.6 0.65
alv:Alvin_2866 hypothetical protein 36.6 3.2
ooe:OEOE_1831 Beta-galactosidase 38.1 7.3
> asn:102382508 endonuclease domain-containing 1 protein-like
Length=540
Score = 41.6 bits (96), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 56/251 (22%), Positives = 109/251 (43%), Gaps = 38/251 (15%)
Query 108 SKVLLPQFRVSTAVASIYEN-DTNRGQVNPSSLLSYLGIKGFGYSKTNSHIRTFPA--IF 164
S + L Q + + A++ YEN + +RGQ+NP+S G + T + T P F
Sbjct 112 SGLTLEQLQSAQAISRDYENTNYDRGQLNPNSFQY-----DSGSTATFTLTNTVPMNHCF 166
Query 165 NLAYWDIFKNYYANKQEEN-------AYVITGINHIWKKISAGDGVAWNREWTEN----S 213
N +W + +K ++N AY++TG +KI D W+R+ T N +
Sbjct 167 NQMHWRKLEESLRDKLKQNCLNEGGTAYLVTGAVAGTEKIPKKDDQEWDRQRTYNRVAVA 226
Query 214 SEAYPLNPSSQAPSFIKLEFEEKISPEEVNEIQFLTNDPEKPSQKINKLTKLGNAFIFER 273
S + A + +K F +E ++++F + ++ ++K+ ++ N +
Sbjct 227 SHIWTAVCCDNANNNLKFSFAILAENKEESKLRFFS--VKQLNKKLQEMYSTQNIRVLA- 283
Query 274 TDPEAMGLKKPDNPKRATNIYVYQVKKPIYLAYNMNIAGNNYLTMPDNKKIKLTPFPLKN 333
D A K T + +K+ +Y A+ I + +P +K+ KN
Sbjct 284 DDCNAQSQK--------TQEVLSAIKQDLYSAFWDLILNLYHQVLPSDKR--------KN 327
Query 334 IDQERNKILAA 344
+D+E K++ +
Sbjct 328 LDKETTKVMKS 338
> alv:Alvin_2866 hypothetical protein
Length=89
Score = 36.6 bits (83), Expect = 3.2, Method: Composition-based stats.
Identities = 30/87 (34%), Positives = 38/87 (44%), Gaps = 4/87 (5%)
Query 492 ATEEEPLGTLAGR-GVATMYKSGRGLKIKCTEPSMIMALG--SITPRIDYSQGNKWWTRL 548
+ + E L T R V M +GRGL+ E A G + S+G KW
Sbjct 2 SDQTETLMTFPCRFAVKAMGPAGRGLEAVVVEIVSRHAEGVDETAISVRESRGGKWIAVT 61
Query 549 QNMDDFHKPTLDAIGFQELIAEEAAAW 575
+ KP LDAI +QEL A E AW
Sbjct 62 LTFEAHSKPQLDAI-YQELSAHELVAW 87
> ooe:OEOE_1831 Beta-galactosidase
Length=671
Score = 38.1 bits (87), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 54/124 (44%), Gaps = 8/124 (6%)
Query 345 PSTSAYVVEGATMPYGAATKTI--GLPSYDRREKYNSSNAWYSQAGLAVKTYLSDRFNNW 402
P+ Y V+ T YG ++ + G Y ++E N + + GL Y S+R N+W
Sbjct 134 PAIIGYQVDNETKSYGTSSSNVQAGFVKYIKKEFNNDIEKFNREFGL---DYWSNRINSW 190
Query 403 LNTEWIDGTTGGINSITAVDVSDGKLTMDALILQKKIFNMLNRVA--ITDGTYQAWREAT 460
+ +DGT G + T +L D L Q I N + +T WR+ +
Sbjct 191 EDFPSMDGTING-SLGTEFAKYQRQLVTDYLKWQVNIVNEYKKSKQFVTHNFDFEWRDYS 249
Query 461 YGIR 464
YGI+
Sbjct 250 YGIQ 253
Lambda K H a alpha
0.316 0.132 0.395 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1617285166872