bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_7
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
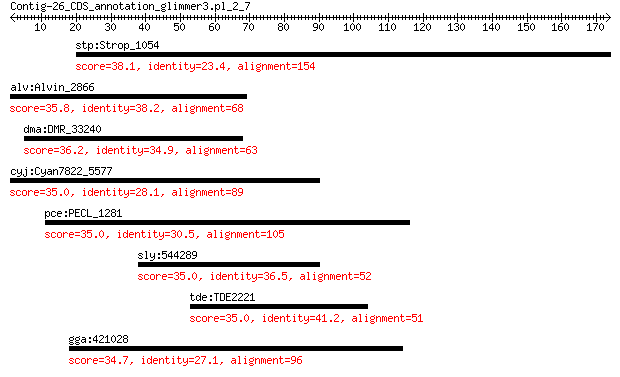
stp:Strop_1054 YD repeat-containing protein 38.1 0.69
alv:Alvin_2866 hypothetical protein 35.8 0.91
dma:DMR_33240 ftsQ; cell division protein FtsQ 36.2 2.4
cyj:Cyan7822_5577 hypothetical protein 35.0 5.1
pce:PECL_1281 rpoA; DNA-directed RNA polymerase subunit alpha 35.0 6.6
sly:544289 PME2.1, PE_2.1; pectin esterase (EC:3.1.1.11) 35.0 6.7
tde:TDE2221 hypothetical protein 35.0 7.3
gga:421028 DSEL; dermatan sulfate epimerase-like 34.7 9.3
> stp:Strop_1054 YD repeat-containing protein
Length=2286
Score = 38.1 bits (87), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 36/158 (23%), Positives = 61/158 (39%), Gaps = 13/158 (8%)
Query 20 ALGSITPRIDYSQGNKWWTRLQNMDDFHKPTLDAIGFQELIAEEAAAWSTEATGNHELVY 79
+G +T Y+ G+ + +R+ D + +PT ++ + +E AA S
Sbjct 1379 GIGKLTAATRYTNGDPYTSRVDAYDAYGRPTSTSVVLPDTQSELCAAASPNTCTYTTTYT 1438
Query 80 QSLGKQPSWIEYTTDVNETYGDFAAGMPLAFMCLNRVYEENSDHTIANASTYIDPTIYNK 139
QP I AA +P + L N ++++A Y + IYNK
Sbjct 1439 YRPNGQPWRIGMPA---------AADLPAETLILGYTDVGNPAGSLSSAQIYANTVIYNK 1489
Query 140 IFAESRLSSQNFWVQVA----FDVTARRVMSAKQIPNL 173
I ++ F +VA D RR+ S +P L
Sbjct 1490 IDQLTQYQLGRFGRRVAVTSSIDEPTRRLTSTNVVPEL 1527
> alv:Alvin_2866 hypothetical protein
Length=89
Score = 35.8 bits (81), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 32/70 (46%), Gaps = 3/70 (4%)
Query 1 MYKSGRGLKIKCTEPCMIMALGSITPRIDY--SQGNKWWTRLQNMDDFHKPTLDAIGFQE 58
M +GRGL+ E A G I S+G KW + KP LDAI +QE
Sbjct 20 MGPAGRGLEAVVVEIVSRHAEGVDETAISVRESRGGKWIAVTLTFEAHSKPQLDAI-YQE 78
Query 59 LIAEEAAAWS 68
L A E AW+
Sbjct 79 LSAHELVAWA 88
> dma:DMR_33240 ftsQ; cell division protein FtsQ
Length=313
Score = 36.2 bits (82), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 22/63 (35%), Positives = 32/63 (51%), Gaps = 7/63 (11%)
Query 5 GRGLKIKCTEPCMIMALGSITPRIDYSQGNKWWTRLQNMDDFHKPTLDAIGFQELIAEEA 64
GRGL+I+ EP +++ LGS R + S+ N W MD + LD +G + AE
Sbjct 245 GRGLEIRLMEPGIVLCLGSQNWRRNLSRMNMVW-----MDLRRRGELDKVGL--ITAEGD 297
Query 65 AAW 67
W
Sbjct 298 KVW 300
> cyj:Cyan7822_5577 hypothetical protein
Length=212
Score = 35.0 bits (79), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 42/96 (44%), Gaps = 15/96 (16%)
Query 1 MYKSGRGLKIKCTEPCMIMALGS--ITPRIDYSQGNK-----WWTRLQNMDDFHKPTLDA 53
++ S RG+ I T+ C G+ + P + Y G+K W T + ++D + P
Sbjct 66 LFASLRGIPINGTDNCTYRKTGTHQMQPDVSYYIGDKANIIPWGTSIVDLDVYPPP---- 121
Query 54 IGFQELIAEEAAAWSTEATGNHELVYQSLGKQPSWI 89
+L+ E A + G L+Y+ L Q WI
Sbjct 122 ----DLVIEVANTSLADDKGEKRLLYEDLNIQEYWI 153
> pce:PECL_1281 rpoA; DNA-directed RNA polymerase subunit alpha
Length=314
Score = 35.0 bits (79), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 32/116 (28%), Positives = 50/116 (43%), Gaps = 14/116 (12%)
Query 11 KCTEPCMIMALGSI-TP--RIDYSQGNKWWTRLQNMDDFHKPTLDAIGFQELIAEEAAAW 67
K P ++ + SI TP R++Y N TR+ DD+ K TLD L A EA +
Sbjct 156 KVDMPIGVLPIDSIYTPIERVNYQVEN---TRVGQRDDYDKLTLDVWTDGSLAASEAISL 212
Query 68 STEATGNHELVYQSLGKQPSWIEYTTDVNETYGDFAAGMPL--------AFMCLNR 115
S + H ++ +L Q E + ET+ + M + ++ CL R
Sbjct 213 SAKILTEHLTIFVNLDDQAKDTEIMVEKEETHKEKMLEMTIEELDLSVRSYNCLKR 268
> sly:544289 PME2.1, PE_2.1; pectin esterase (EC:3.1.1.11)
Length=550
Score = 35.0 bits (79), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 19/55 (35%), Positives = 27/55 (49%), Gaps = 3/55 (5%)
Query 38 TRLQNMDDFHKPTLDAIGFQELIAEEAAAWSTEA---TGNHELVYQSLGKQPSWI 89
T L +D F K ++ ELI+ A + A T N E++ LGK PSW+
Sbjct 165 TCLDELDSFTKAMINGTNLDELISRAKVALAMLASVTTPNDEVLRPGLGKMPSWV 219
> tde:TDE2221 hypothetical protein
Length=344
Score = 35.0 bits (79), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 31/52 (60%), Gaps = 3/52 (6%)
Query 53 AIGFQELIAEEAAAWSTEATGNHELVYQSLG-KQPSWIEYTTDVNETYGDFA 103
A GF+ LIAEE +AW +G++ L+YQ L K+P+W + + T G F
Sbjct 17 AEGFK-LIAEEKSAWEGTESGDY-LIYQDLSWKEPTWAGFLYYNDNTIGSFV 66
> gga:421028 DSEL; dermatan sulfate epimerase-like
Length=1216
Score = 34.7 bits (78), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 26/122 (21%), Positives = 49/122 (40%), Gaps = 28/122 (23%)
Query 18 IMALGSITPRIDYSQGNKWWTRLQNMDDFHKPTLDAIGFQELI------------AEEAA 65
+M T S + W T ++N+D + K LD+ G ++ + AE+
Sbjct 14 LMMFAVFTFEESVSNYSDWVTFMENLDQYDKEKLDSFGAEQKVKKVSLHPSLYFDAEDVH 73
Query 66 AWSTEATGNHELVYQSLGKQ--------------PSWIEYTTDVNETYGDFAAGMPLAFM 111
+A +H +++++ P ++++ NE YG+ PLAF
Sbjct 74 VLRQKAHTSHSHIFRTIRSAVMVMLSNPMYYLPPPKYVDFAAKWNEVYGNNLP--PLAFY 131
Query 112 CL 113
CL
Sbjct 132 CL 133
Lambda K H a alpha
0.318 0.131 0.405 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 144537900232