bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-27_CDS_annotation_glimmer3.pl_2_2
Length=388
Score E
Sequences producing significant alignments: (Bits) Value
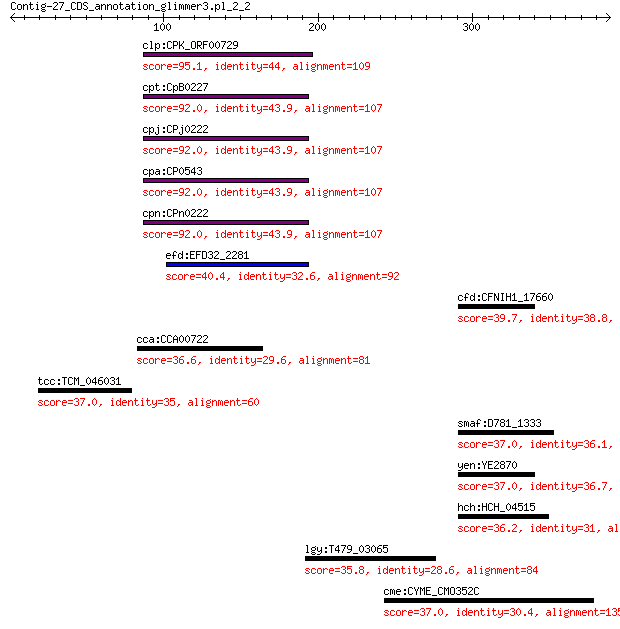
clp:CPK_ORF00729 hypothetical protein 95.1 6e-21
cpt:CpB0227 hypothetical protein 92.0 8e-20
cpj:CPj0222 hypothetical protein 92.0 8e-20
cpa:CP0543 hypothetical protein 92.0 8e-20
cpn:CPn0222 hypothetical protein 92.0 8e-20
efd:EFD32_2281 hypothetical protein 40.4 0.47
cfd:CFNIH1_17660 urea ABC transporter ATP-binding protein 39.7 0.70
cca:CCA00722 hypothetical protein 36.6 1.8
tcc:TCM_046031 hypothetical protein 37.0 2.7
smaf:D781_1333 urea ABC transporter, ATP-binding protein UrtD 37.0 5.2
yen:YE2870 ABC transport ATP-binding subunit 37.0 5.4
hch:HCH_04515 putative ABC transporter ATPase 36.2 8.1
lgy:T479_03065 Parasporal protein 35.8 9.5
cme:CYME_CMO352C ATP-dependent RNA helicase 37.0 9.5
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 95.1 bits (235), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 48/109 (44%), Positives = 61/109 (56%), Gaps = 13/109 (12%)
Query 87 WVEIPCGKCEGCRIARSREWANRCMMELEYHDSAYFLTLTYDEEHVPRHWYADPETGEAM 146
WV +PC KC CR ++ W+ RC+ E ++ FLTLTYD++H+P
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLP------------- 65
Query 147 QSLSLCKRDLQLFWKRLRKAFPDDHIRYFACGEYGSTTFRPHYHAIVFG 195
Q SL K LQLF KRLR IRYF CGEYG+ RPHYH ++F
Sbjct 66 QYGSLVKLHLQLFLKRLRDRISPHKIRYFGCGEYGTKLQRPHYHLLIFN 114
> cpt:CpB0227 hypothetical protein
Length=113
Score = 92.0 bits (227), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 47/107 (44%), Positives = 60/107 (56%), Gaps = 13/107 (12%)
Query 87 WVEIPCGKCEGCRIARSREWANRCMMELEYHDSAYFLTLTYDEEHVPRHWYADPETGEAM 146
WV +PC KC CR ++ W+ RC+ E ++ FLTLTYD++H+P
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLP------------- 65
Query 147 QSLSLCKRDLQLFWKRLRKAFPDDHIRYFACGEYGSTTFRPHYHAIV 193
Q SL K LQLF KRLRK IRYF CG YG+ RPHYH ++
Sbjct 66 QYGSLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpj:CPj0222 hypothetical protein
Length=113
Score = 92.0 bits (227), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 47/107 (44%), Positives = 60/107 (56%), Gaps = 13/107 (12%)
Query 87 WVEIPCGKCEGCRIARSREWANRCMMELEYHDSAYFLTLTYDEEHVPRHWYADPETGEAM 146
WV +PC KC CR ++ W+ RC+ E ++ FLTLTYD++H+P
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLP------------- 65
Query 147 QSLSLCKRDLQLFWKRLRKAFPDDHIRYFACGEYGSTTFRPHYHAIV 193
Q SL K LQLF KRLRK IRYF CG YG+ RPHYH ++
Sbjct 66 QYGSLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpa:CP0543 hypothetical protein
Length=113
Score = 92.0 bits (227), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 47/107 (44%), Positives = 60/107 (56%), Gaps = 13/107 (12%)
Query 87 WVEIPCGKCEGCRIARSREWANRCMMELEYHDSAYFLTLTYDEEHVPRHWYADPETGEAM 146
WV +PC KC CR ++ W+ RC+ E ++ FLTLTYD++H+P
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLP------------- 65
Query 147 QSLSLCKRDLQLFWKRLRKAFPDDHIRYFACGEYGSTTFRPHYHAIV 193
Q SL K LQLF KRLRK IRYF CG YG+ RPHYH ++
Sbjct 66 QYGSLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> cpn:CPn0222 hypothetical protein
Length=113
Score = 92.0 bits (227), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 47/107 (44%), Positives = 60/107 (56%), Gaps = 13/107 (12%)
Query 87 WVEIPCGKCEGCRIARSREWANRCMMELEYHDSAYFLTLTYDEEHVPRHWYADPETGEAM 146
WV +PC KC CR ++ W+ RC+ E ++ FLTLTYD++H+P
Sbjct 19 WVLMPCLKCRFCRTQHAKVWSYRCVHEASLYEKNCFLTLTYDDKHLP------------- 65
Query 147 QSLSLCKRDLQLFWKRLRKAFPDDHIRYFACGEYGSTTFRPHYHAIV 193
Q SL K LQLF KRLRK IRYF CG YG+ RPHYH ++
Sbjct 66 QYGSLVKLHLQLFLKRLRKMISPHKIRYFECGAYGTKLQRPHYHLLL 112
> efd:EFD32_2281 hypothetical protein
Length=271
Score = 40.4 bits (93), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 45/99 (45%), Gaps = 19/99 (19%)
Query 102 RSREWANRCMMELEYHDSAYFLTLTYDEEHVPRHWYADPETGEAMQSLSLCKRDLQLFWK 161
RS +W R M + Y+LTLTYDE +P PE E K+DL F +
Sbjct 52 RSYKWM-RLAMNGNFFKGDYYLTLTYDEGDIP-----PPEKAEE------AKKDLSNFLR 99
Query 162 RLRKAFP--DDHIRYFACGEY-----GSTTFRPHYHAIV 193
++R + D ++Y EY G+ R H+H ++
Sbjct 100 KVRNLYKKVDKELKYIWVMEYELDQEGNYLKRVHFHLVM 138
> cfd:CFNIH1_17660 urea ABC transporter ATP-binding protein
Length=242
Score = 39.7 bits (91), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 19/49 (39%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 291 RPGIGRQWYDDHPECMEYDTISISTPDGGRKIRPPKYFDKLFDLEQPEL 339
RP GR YD + M +D+I+I+ GRK + P F+ L E EL
Sbjct 52 RPQSGRAIYDQSIDLMTFDSIAIARQGIGRKFQKPTVFEALTVAENLEL 100
> cca:CCA00722 hypothetical protein
Length=117
Score = 36.6 bits (83), Expect = 1.8, Method: Composition-based stats.
Identities = 24/81 (30%), Positives = 38/81 (47%), Gaps = 13/81 (16%)
Query 83 AWLDWVEIPCGKCEGCRIARSREWANRCMMELEYHDSAYFLTLTYDEEHVPRHWYADPET 142
A + + P K E ++ W+ RC+ E + FLTLTY++ ++ PE
Sbjct 23 ARITYFRTPMHKNEVLVHGSAKVWSYRCIHEASLYGQNSFLTLTYEDRNL-------PEK 75
Query 143 GEAMQSLSLCKRDLQLFWKRL 163
G SL +RD++LF R
Sbjct 76 G------SLVRRDVRLFLMRF 90
> tcc:TCM_046031 hypothetical protein
Length=148
Score = 37.0 bits (84), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/64 (33%), Positives = 33/64 (52%), Gaps = 4/64 (6%)
Query 19 TRPTEYGEG---DFIACFHPLKGFRIGTTKNGKADMKIVPYGVHHLELRKGR-ICTSDVP 74
+ P EYG G I HP+ F + +T + + D++++PY H + R R I S VP
Sbjct 28 SEPREYGPGITTSRIPTDHPIIAFSLLSTPHRRWDLRLLPYAFHRIWCRSTRLILLSPVP 87
Query 75 EISA 78
S+
Sbjct 88 ASSS 91
> smaf:D781_1333 urea ABC transporter, ATP-binding protein UrtD
Length=268
Score = 37.0 bits (84), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 31/61 (51%), Gaps = 2/61 (3%)
Query 291 RPGIGRQWYDDHPECMEYDTISISTPDGGRKIRPPKYFDKLFDLEQPELMAAIKEKRKHF 350
RP GR +YD + M+ D I I+ GRK + P F+ L E E+ A K +R +
Sbjct 78 RPDSGRVFYDQTRDLMQMDPIRIAQAGIGRKFQKPTVFEALTVFENLEI--AQKNRRSVW 135
Query 351 A 351
A
Sbjct 136 A 136
> yen:YE2870 ABC transport ATP-binding subunit
Length=278
Score = 37.0 bits (84), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 18/49 (37%), Positives = 24/49 (49%), Gaps = 0/49 (0%)
Query 291 RPGIGRQWYDDHPECMEYDTISISTPDGGRKIRPPKYFDKLFDLEQPEL 339
RP G+ YD H + +TI I+ GRK + P F+ L E EL
Sbjct 88 RPQSGKMMYDQHHDLSVMNTIEIAQAGIGRKFQKPTVFEALSVFENLEL 136
> hch:HCH_04515 putative ABC transporter ATPase
Length=252
Score = 36.2 bits (82), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 31/58 (53%), Gaps = 2/58 (3%)
Query 291 RPGIGRQWYDDHPECMEYDTISISTPDGGRKIRPPKYFDKLFDLEQPELMAAIKEKRK 348
RP +G+ +++H + ++D I+ GRK + P + L LE EL A++ RK
Sbjct 62 RPDVGQVLFNNHIDLTQHDEAEIANMGVGRKFQKPTVIESLTVLENLEL--ALQGGRK 117
> lgy:T479_03065 Parasporal protein
Length=219
Score = 35.8 bits (81), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 24/86 (28%), Positives = 39/86 (45%), Gaps = 13/86 (15%)
Query 192 IVFGLHLHDLMPVQDIRRGDVGYQYFYSEALQKAWSVVEQKGEYDTPCTRKPIGYVLVGQ 251
IVFG P Q + RG+ Y F ++ALQ + V+ G D P + + GY+
Sbjct 58 IVFGTTPSTFSPYQQLTRGEAAY--FLAQALQLQTNNVDNPGFSDVPTSHQYYGYI---- 111
Query 252 VNWETCAYVARYVLKKAS--GPEADV 275
A A +++K + P+A +
Sbjct 112 -----AALAANGIIQKGNQFNPDASI 132
> cme:CYME_CMO352C ATP-dependent RNA helicase
Length=1214
Score = 37.0 bits (84), Expect = 9.5, Method: Composition-based stats.
Identities = 41/141 (29%), Positives = 66/141 (47%), Gaps = 23/141 (16%)
Query 243 PIGYVLVGQVNWETCAYVARYVLKKASGPEADVYQTFNIDPEY---VDMSRRPGIGRQWY 299
PIGYV+V Q WE A VA +L++ AD ++ N P Y +++ R PG Q
Sbjct 576 PIGYVVVMQSAWEPSAEVACQLLQRG----ADALRS-NFVPSYGMALNLLRYPGTPLQSA 630
Query 300 DDHPECMEYDTISISTPDGGRKIRPPKYFDKLFDLEQPELMAAIKEKRKHFAEEGKKAKL 359
+ +E S G P+ D++ LE+ +KE + FA+ G + +
Sbjct 631 RRY---LERSFGSFLASRGRLNQWKPEIHDEVEALER-----QVKEAHRIFAQHGGEKVV 682
Query 360 AQSTMTYEEILE---TQERVL 377
A Y+++LE +ER+L
Sbjct 683 A----AYDKLLERLRCEERIL 699
Lambda K H a alpha
0.321 0.139 0.444 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 769174474406