bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-29_CDS_annotation_glimmer3.pl_2_5
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
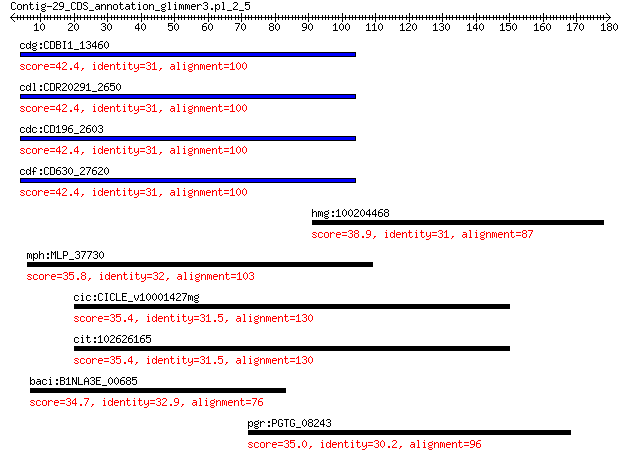
cdg:CDBI1_13460 UDP pyrophosphate synthetase 42.4 0.019
cdl:CDR20291_2650 uppS; UDP pyrophosphate synthetase 42.4 0.019
cdc:CD196_2603 uppS; UDP pyrophosphate synthetase 42.4 0.019
cdf:CD630_27620 uppS; undecaprenyl pyrophosphate synthetase (E... 42.4 0.019
hmg:100204468 hexaprenyldihydroxybenzoate methyltransferase, m... 38.9 0.27
mph:MLP_37730 copper-transporting ATPase (EC:3.6.3.4) 35.8 4.5
cic:CICLE_v10001427mg hypothetical protein 35.4 5.6
cit:102626165 uncharacterized LOC102626165 35.4 5.6
baci:B1NLA3E_00685 cbiO; cobalt transporter ATP-binding subunit 34.7 7.5
pgr:PGTG_08243 hypothetical protein 35.0 9.2
> cdg:CDBI1_13460 UDP pyrophosphate synthetase
Length=214
Score = 42.4 bits (98), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 31/104 (30%), Positives = 44/104 (42%), Gaps = 5/104 (5%)
Query 4 SILSARRIKMPKFANQFEPHA-RIHTNPGTPEKVL---YGPVFDENGTLDLEPKGKENLY 59
++L K P F + P+ RI G + L YG +D N +E K N+
Sbjct 88 ALLVVGNTKSPMFPEELLPYTNRIKAKEGQIKANLLVNYGWYWDLNNISGVENVNKNNIQ 147
Query 60 DYIQSHKDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLPRTY 103
DYIQS D + LI+ R LS Q +Y DF + +
Sbjct 148 DYIQSF-DVSRLDLIIRWGGRRRLSGFLPVQSIYADFYVIDNYW 190
> cdl:CDR20291_2650 uppS; UDP pyrophosphate synthetase
Length=214
Score = 42.4 bits (98), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 31/104 (30%), Positives = 44/104 (42%), Gaps = 5/104 (5%)
Query 4 SILSARRIKMPKFANQFEPHA-RIHTNPGTPEKVL---YGPVFDENGTLDLEPKGKENLY 59
++L K P F + P+ RI G + L YG +D N +E K N+
Sbjct 88 ALLVVGNTKSPMFPEELLPYTNRIKAKEGQIKANLLVNYGWYWDLNNISGVENVNKNNIQ 147
Query 60 DYIQSHKDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLPRTY 103
DYIQS D + LI+ R LS Q +Y DF + +
Sbjct 148 DYIQSF-DVSRLDLIIRWGGRRRLSGFLPVQSIYADFYVIDNYW 190
> cdc:CD196_2603 uppS; UDP pyrophosphate synthetase
Length=214
Score = 42.4 bits (98), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 31/104 (30%), Positives = 44/104 (42%), Gaps = 5/104 (5%)
Query 4 SILSARRIKMPKFANQFEPHA-RIHTNPGTPEKVL---YGPVFDENGTLDLEPKGKENLY 59
++L K P F + P+ RI G + L YG +D N +E K N+
Sbjct 88 ALLVVGNTKSPMFPEELLPYTNRIKAKEGQIKANLLVNYGWYWDLNNISGVENVNKNNIQ 147
Query 60 DYIQSHKDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLPRTY 103
DYIQS D + LI+ R LS Q +Y DF + +
Sbjct 148 DYIQSF-DVSRLDLIIRWGGRRRLSGFLPVQSIYADFYVIDNYW 190
> cdf:CD630_27620 uppS; undecaprenyl pyrophosphate synthetase
(EC:2.5.1.31)
Length=214
Score = 42.4 bits (98), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 31/104 (30%), Positives = 44/104 (42%), Gaps = 5/104 (5%)
Query 4 SILSARRIKMPKFANQFEPHA-RIHTNPGTPEKVL---YGPVFDENGTLDLEPKGKENLY 59
++L K P F + P+ RI G + L YG +D N +E K N+
Sbjct 88 ALLVVGNTKSPMFPEELLPYTNRIKAKEGQIKANLLVNYGWYWDLNNISGVENVNKNNIQ 147
Query 60 DYIQSHKDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLPRTY 103
DYIQS D + LI+ R LS Q +Y DF + +
Sbjct 148 DYIQSF-DVSRLDLIIRWGGRRRLSGFLPVQSIYADFYVIDNYW 190
> hmg:100204468 hexaprenyldihydroxybenzoate methyltransferase,
mitochondrial-like
Length=235
Score = 38.9 bits (89), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 43/87 (49%), Gaps = 6/87 (7%)
Query 91 GMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAKFDHDPHKFIVSMDKPGFLEKLGV 150
G Y FTT+ RT+ +L A AE+ F +P+ T HD +KF+ + G+L+ G+
Sbjct 145 GGYILFTTINRTFIASLLAKYVAEYIFRIVPINT-----HDENKFVPPYELTGYLQNSGM 199
Query 151 PVPNSGAEQQSPTLTTSAATAPAGEVS 177
P +P LT + P+ V+
Sbjct 200 YKPTYLGMSLNP-LTMQWSWIPSTLVN 225
> mph:MLP_37730 copper-transporting ATPase (EC:3.6.3.4)
Length=839
Score = 35.8 bits (81), Expect = 4.5, Method: Composition-based stats.
Identities = 33/103 (32%), Positives = 43/103 (42%), Gaps = 14/103 (14%)
Query 6 LSARRIKMPKFANQFEPHARIHTNPGTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSH 65
L A+R + K A E ARI T VFD+ GTL KG+ + DY+
Sbjct 475 LGAKRGVLFKNATGIETAARIDT-----------VVFDKTGTLT---KGEPEVTDYLAVG 520
Query 66 KDSCDIKLIVDRCARGDLSALSKAQGMYGDFTTLPRTYAEALQ 108
D D+ V R L+KA Y D +PR A A +
Sbjct 521 TDDLDLLSFVVAAERESEHPLAKAIVAYADARDIPRRTATAFR 563
> cic:CICLE_v10001427mg hypothetical protein
Length=392
Score = 35.4 bits (80), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 41/147 (28%), Positives = 59/147 (40%), Gaps = 21/147 (14%)
Query 20 FEPHARI---HTNP-------GTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDS- 68
E HARI HTN G +KV+Y V E G ++ + ++S K
Sbjct 147 IEGHARIKRSHTNSDKEEDAAGENKKVIYEDVSMERGAFLVQQAMRAFRAQNVESAKSRL 206
Query 69 --C--DIKLIVDRCARGD--LSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPV 122
C DI+ ++R S L GM GD A+ AD+ F M+LP+
Sbjct 207 SLCTEDIRDQIERMGNTSELCSQLGAVLGMLGDCCRAMGDADAAVAYFADSVEFLMKLPM 266
Query 123 ETRAKFDHDPHKFIVSMDKPGFLEKLG 149
+ H VS++K G L+ G
Sbjct 267 DDLEII----HTLSVSLNKIGDLKYYG 289
> cit:102626165 uncharacterized LOC102626165
Length=392
Score = 35.4 bits (80), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 41/147 (28%), Positives = 59/147 (40%), Gaps = 21/147 (14%)
Query 20 FEPHARI---HTNP-------GTPEKVLYGPVFDENGTLDLEPKGKENLYDYIQSHKDS- 68
E HARI HTN G +KV+Y V E G ++ + ++S K
Sbjct 147 IEGHARIKRSHTNSDKEEDAAGENKKVIYEDVSMERGAFLVQQAMRAFRAQNVESAKSRL 206
Query 69 --C--DIKLIVDRCARGD--LSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPV 122
C DI+ ++R S L GM GD A+ AD+ F M+LP+
Sbjct 207 SLCTEDIRDQIERMGNTSELCSQLGAVLGMLGDCCRAMGDADAAVAYFADSVEFLMKLPM 266
Query 123 ETRAKFDHDPHKFIVSMDKPGFLEKLG 149
+ H VS++K G L+ G
Sbjct 267 DDLEII----HTLSVSLNKIGDLKYYG 289
> baci:B1NLA3E_00685 cbiO; cobalt transporter ATP-binding subunit
Length=279
Score = 34.7 bits (78), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 25/90 (28%), Positives = 47/90 (52%), Gaps = 19/90 (21%)
Query 7 SARRIKMPKFANQFEPHARIHTNPGTPEKVLYGPVF---------DENGTLDLEPKGKEN 57
S ++++M +F +Q EPH H + G ++V V DE ++ L+P+G+E
Sbjct 124 SLKKVEMDQFLDQ-EPH---HLSGGQKQRVAIASVLALQPAIIILDEATSM-LDPRGREE 178
Query 58 LYDYIQSHKDSCDIKLI-----VDRCARGD 82
+ + ++ KDS +I +I ++ AR D
Sbjct 179 VLNIVRELKDSRNITVISITHDLEEAARAD 208
> pgr:PGTG_08243 hypothetical protein
Length=1917
Score = 35.0 bits (79), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 42/100 (42%), Gaps = 8/100 (8%)
Query 72 KLIVDRCAR----GDLSALSKAQGMYGDFTTLPRTYAEALQALADAEHFFMRLPVETRAK 127
KL R +R GD S L DF P T++ L A P T +
Sbjct 24 KLTRSRASRPPSIGDASQLFGGGNGLDDFFGPPSTFSNDLAQQASLFDPVPEEPANTSSS 83
Query 128 FDHDPHKFIVSMDKPGFLEKLGVPVPNSGAEQQSPTLTTS 167
FDHDP + S D+P + P+PN+ ++ P ++ S
Sbjct 84 FDHDPFAW-PSNDQPA---QHPSPIPNNSSQISDPNISAS 119
Lambda K H a alpha
0.317 0.134 0.395 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 160264878837