bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_7
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
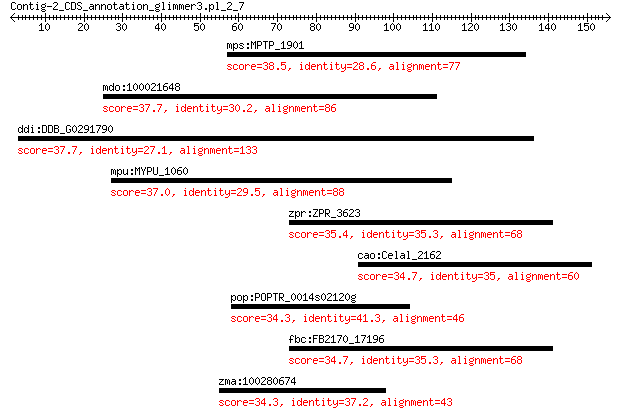
mps:MPTP_1901 hypothetical protein 38.5 0.064
mdo:100021648 FAN1; FANCD2/FANCI-associated nuclease 1 37.7
ddi:DDB_G0291790 hypothetical protein 37.7 0.65
mpu:MYPU_1060 hemK; protoporphirogen oxidase HemK 37.0 0.91
zpr:ZPR_3623 aspartokinase 35.4 4.6
cao:Celal_2162 aspartate kinase (EC:1.1.1.3 2.7.2.4) 34.7 7.8
pop:POPTR_0014s02120g cyclin A3.1 family protein 34.3 7.8
fbc:FB2170_17196 bifunctional aspartokinase I/homoserine dehyd... 34.7 8.4
zma:100280674 cyclin-A2 34.3 9.1
> mps:MPTP_1901 hypothetical protein
Length=95
Score = 38.5 bits (88), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 22/83 (27%), Positives = 39/83 (47%), Gaps = 6/83 (7%)
Query 57 YNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFY------LNRWFASGGSSQASISWPFF 110
Y RY I+ + L T+ D V P+ DF+ LN + ++ ++ PF
Sbjct 10 YVERYNEETPVIEIMQDKKLVTMIDEVDPVLDFFRNDPNALNGGLGTMSLAKLNLLLPFI 69
Query 111 KVNPNTLDSIFAVAADSTWESDQ 133
++P TLD I A+ ++ S++
Sbjct 70 TISPETLDQITAILCETPILSER 92
> mdo:100021648 FAN1; FANCD2/FANCI-associated nuclease 1
Length=1061
Score = 37.7 bits (86), Expect = 0.62, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 42/86 (49%), Gaps = 10/86 (12%)
Query 25 FDNIGMEAVPAITLFNSNAFDNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVA 84
+D I M+ +P + + AF DL +D F Y R +SK+ ++H A TLK+W+A
Sbjct 891 WDIIFMDGIPDVFRNSYQAFPLDLYTD-SF--YENRRNAIESKLQKIHEASPETLKEWIA 947
Query 85 PIDDFYLNRWFASGGSSQASISWPFF 110
+ W A G + ++W F
Sbjct 948 DV-------WNAHEGEVASGVTWDRF 966
> ddi:DDB_G0291790 hypothetical protein
Length=391
Score = 37.7 bits (86), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 36/138 (26%), Positives = 61/138 (44%), Gaps = 18/138 (13%)
Query 3 YAISGQDSQLLCTSVEDLPIPEFDNIGMEAVPAITLFNSNAFDNDLESDFDFLGYNPRYW 62
++ S +D+ +L +DL + + + + AI +F + DN Y R+
Sbjct 87 FSKSEKDTTILV-GYQDLIFKQMKKLDPKEIAAIIIFYNLIIDNT--------NYQGRFG 137
Query 63 PWKSKIDRVHGAFLTTLKDWVAP--IDDFYLNRWFASGGSSQASISWPFFKVNPNTLD-- 118
K + R+HG FL TL + IDD L ++F S + S+ W F N
Sbjct 138 INKQFLSRLHGEFLRTLPKEIIKVIIDD--LVKYFLSTNYYEKSLYW--FNANSTIFSNE 193
Query 119 -SIFAVAADSTWESDQLL 135
S A+ + T+++ QLL
Sbjct 194 ISKTAIFSFITYQNSQLL 211
> mpu:MYPU_1060 hemK; protoporphirogen oxidase HemK
Length=230
Score = 37.0 bits (84), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 41/89 (46%), Gaps = 3/89 (3%)
Query 27 NIGMEAVPAITLFNSNAFDNDLESDFDFLGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPI 86
N+ + V + S+ F+N ++ DFD + NP Y ++ KID+ F LK AP
Sbjct 116 NVKINNVKNYKIIQSDLFEN-IQGDFDIIVSNPPYLSYEQKIDK-SVKFFEPLKALYAPK 173
Query 87 DDFYLNRWFASGGSS-QASISWPFFKVNP 114
+ +Y SS FF++NP
Sbjct 174 NGWYFYEKIIEKASSFLKKDGMLFFEINP 202
> zpr:ZPR_3623 aspartokinase
Length=1088
Score = 35.4 bits (80), Expect = 4.6, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 37/79 (47%), Gaps = 13/79 (16%)
Query 73 GAFLTTLKDWVAP---------IDDFYLNRWFA-SGGSSQASISWPFFKVNPNTLDSIFA 122
G + KD+VA +D ++ FA GGS I+W ++P D++
Sbjct 48 GILIENYKDFVARDIARIFLLGLDSLKIDHLFALVGGSLGGGIAWEMAALSPEITDNLIT 107
Query 123 VAADSTWES-DQLLINCDV 140
VA D W+S D L+ NC +
Sbjct 108 VATD--WKSTDWLIANCQI 124
> cao:Celal_2162 aspartate kinase (EC:1.1.1.3 2.7.2.4)
Length=1128
Score = 34.7 bits (78), Expect = 7.8, Method: Composition-based stats.
Identities = 21/62 (34%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Query 91 LNRWFA-SGGSSQASISWPFFKVNPNTLDSIFAVAADSTWES-DQLLINCDVSCKVVRPL 148
+ R FA GGS I+W + PN +++ VA+D W+S D L+ NC + + +
Sbjct 112 VKRLFALIGGSMGGGIAWEMIALEPNLTENLIPVASD--WKSTDWLIANCQIQEQFLLNS 169
Query 149 SQ 150
SQ
Sbjct 170 SQ 171
> pop:POPTR_0014s02120g cyclin A3.1 family protein
Length=363
Score = 34.3 bits (77), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 23/46 (50%), Gaps = 3/46 (7%)
Query 58 NPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFASGGSSQA 103
P+ PW S + + G T LKD V I D YL+R GG QA
Sbjct 291 RPKTHPWSSTLQQYTGYKATDLKDCVLIIHDLYLSR---RGGGLQA 333
> fbc:FB2170_17196 bifunctional aspartokinase I/homoserine dehydrogenase
I
Length=1128
Score = 34.7 bits (78), Expect = 8.4, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 38/79 (48%), Gaps = 13/79 (16%)
Query 73 GAFLTTLKDWVAP---------IDDFYLNRWFAS-GGSSQASISWPFFKVNPNTLDSIFA 122
G + KD+VA + + +++ FA GGS I+W +NPN +
Sbjct 85 GFVIENYKDFVAGDVARIFLLGLKELKIDKLFAMIGGSLGGGIAWEMAALNPNLTTHLIP 144
Query 123 VAADSTWES-DQLLINCDV 140
VA+D W+S D L+ NC +
Sbjct 145 VASD--WKSTDWLIANCQI 161
> zma:100280674 cyclin-A2
Length=423
Score = 34.3 bits (77), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 55 LGYNPRYWPWKSKIDRVHGAFLTTLKDWVAPIDDFYLNRWFAS 97
L +P PW K+ + G ++ LKD + I D LNR F S
Sbjct 346 LTIDPNANPWNMKLQKTTGYKVSELKDCIVAIRDLQLNRKFPS 388
Lambda K H a alpha
0.320 0.137 0.444 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127339103541