bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_1
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
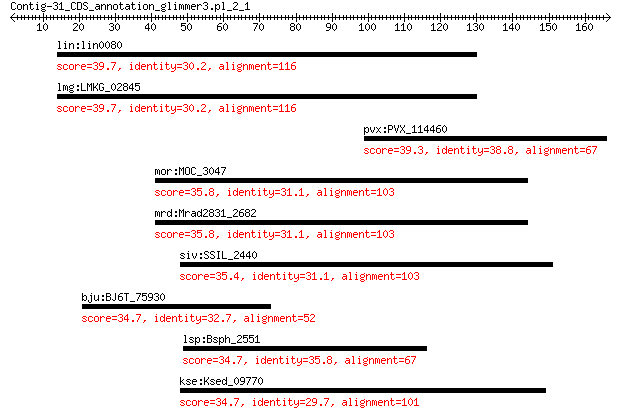
lin:lin0080 hypothetical protein 39.7 0.15
lmg:LMKG_02845 gp36 protein 39.7 0.15
pvx:PVX_114460 hypothetical protein 39.3 0.30
mor:MOC_3047 Short-chain dehydrogenase/reductase SDR (EC:1.3.1.9) 35.8 3.0
mrd:Mrad2831_2682 short-chain dehydrogenase/reductase SDR 35.8 3.1
siv:SSIL_2440 SAM-dependent methyltransferase 35.4 4.3
bju:BJ6T_75930 hypothetical protein 34.7 7.7
lsp:Bsph_2551 hypothetical protein 34.7 8.1
kse:Ksed_09770 NAD-dependent aldehyde dehydrogenase 34.7 9.4
> lin:lin0080 hypothetical protein
Length=257
Score = 39.7 bits (91), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 35/118 (30%), Positives = 51/118 (43%), Gaps = 12/118 (10%)
Query 14 LGFSSDLPSKVQEQFADACQTDTI--IRKYNMMGVNPFIASGGSQYLDTTQIPDFICASN 71
L F S L S E+F D ++ + +RK+ N I + PDF+
Sbjct 77 LIFKSKLES--AERFQDWVTSEVLPSVRKHGAYMTNDTIEKAITD-------PDFLIKLA 127
Query 72 AQVKVKEYFEGLPADIRLEFNNDPMQFAEVVSDPRNTDYLRDIGVLAPLPAEQEGEKH 129
+K +E + + A+ RLE + FAE VSD R T +RD+ L GEK
Sbjct 128 TNLK-EEKTKRIEAEQRLEIQKPKVMFAEAVSDARGTILIRDLAKLIQQNGIDIGEKR 184
> lmg:LMKG_02845 gp36 protein
Length=257
Score = 39.7 bits (91), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 35/118 (30%), Positives = 51/118 (43%), Gaps = 12/118 (10%)
Query 14 LGFSSDLPSKVQEQFADACQTDTI--IRKYNMMGVNPFIASGGSQYLDTTQIPDFICASN 71
L F S L S E+F D ++ + +RK+ N I + PDF+
Sbjct 77 LIFKSKLES--AERFQDWVTSEVLPSVRKHGAYMTNDTIEKAITD-------PDFLIKLA 127
Query 72 AQVKVKEYFEGLPADIRLEFNNDPMQFAEVVSDPRNTDYLRDIGVLAPLPAEQEGEKH 129
+K +E + + A+ RLE + FAE VSD R T +RD+ L GEK
Sbjct 128 TNLK-EEKTKRIEAEQRLEIQKPKVMFAEAVSDARGTILIRDLAKLIQQNGIDIGEKR 184
> pvx:PVX_114460 hypothetical protein
Length=1847
Score = 39.3 bits (90), Expect = 0.30, Method: Composition-based stats.
Identities = 26/67 (39%), Positives = 36/67 (54%), Gaps = 1/67 (1%)
Query 99 AEVVSDPRNTDYLRDIGVLAPLPAEQEGEKHSAQSGDNSKKAPQPSEGSDLFAGKEPEKA 158
A+V S +T + D + A PA E EK SA G +S + P +GS L P+KA
Sbjct 1300 AQVHSTQVHTAQVNDASLGASPPAATEVEK-SAPKGGSSPEGGSPEKGSSLDKANSPDKA 1358
Query 159 VSPEKSN 165
SP+K+N
Sbjct 1359 NSPDKAN 1365
> mor:MOC_3047 Short-chain dehydrogenase/reductase SDR (EC:1.3.1.9)
Length=276
Score = 35.8 bits (81), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 43/109 (39%), Gaps = 11/109 (10%)
Query 41 YNMMGVNPFIASGGSQYLDTTQIPDFICASNAQVKVKEYFEGLP-ADIRLEFNNDPMQFA 99
YN+MGV +YL + PD I + G AD RL FN+
Sbjct 158 YNVMGVAKAALEASVRYLASDLGPDGIRVNALSAGPMRTLAGAGIADARLMFNHQRAH-- 215
Query 100 EVVSDPRNTDYLRDIG-----VLAPLPAEQEGEKHSAQSGDNSKKAPQP 143
+ R T L D+G +L+PL GE H +G N P+P
Sbjct 216 ---APLRRTVSLEDVGGSALYLLSPLSGGVTGEVHFVDAGYNIISMPRP 261
> mrd:Mrad2831_2682 short-chain dehydrogenase/reductase SDR
Length=276
Score = 35.8 bits (81), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 43/109 (39%), Gaps = 11/109 (10%)
Query 41 YNMMGVNPFIASGGSQYLDTTQIPDFICASNAQVKVKEYFEGLP-ADIRLEFNNDPMQFA 99
YN+MGV +YL + PD I + G AD RL FN+
Sbjct 158 YNVMGVAKAALEASVRYLASDLGPDGIRVNALSAGPMRTLAGAGIADARLMFNHQRAH-- 215
Query 100 EVVSDPRNTDYLRDIG-----VLAPLPAEQEGEKHSAQSGDNSKKAPQP 143
+ R T L D+G +L+PL GE H +G N P+P
Sbjct 216 ---APLRRTVSLEDVGGSALYLLSPLSGGVTGEVHFVDAGYNIISMPRP 261
> siv:SSIL_2440 SAM-dependent methyltransferase
Length=262
Score = 35.4 bits (80), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 42/106 (40%), Gaps = 7/106 (7%)
Query 48 PFIASGGSQYLDTTQIPDFICASNAQV---KVKEYFEGLPADIRLEFNNDPMQFAEVVSD 104
FI G Q DTT++P C N QV EY + D DPM F E++ +
Sbjct 138 AFIVKTGMQNYDTTELPLTACMRNIQVVHTTALEYLKNQEDDSFDVVYMDPM-FEEIIEE 196
Query 105 PRNTDYLRDIGVLAPLPAEQEGEKHSAQSGDNSKKAPQPSEGSDLF 150
N + LR G L E E A+ S+ + SD F
Sbjct 197 STNFEALRYAGKHVTLTDEWVAE---AKRVAKSRVVLKAHYKSDWF 239
> bju:BJ6T_75930 hypothetical protein
Length=401
Score = 34.7 bits (78), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 17/52 (33%), Positives = 29/52 (56%), Gaps = 2/52 (4%)
Query 21 PSKVQEQFADACQTDTIIRKYNMMGVNPFIASGGSQYLDTTQIPDFICASNA 72
PSK EQ ++D ++ ++M+G P IA+ +YL+ +IP +S A
Sbjct 88 PSKTVEQVRRLAESDEVLAMFSMLGTGPNIAA--QKYLNAKKIPQLFPSSGA 137
> lsp:Bsph_2551 hypothetical protein
Length=262
Score = 34.7 bits (78), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 33/70 (47%), Gaps = 4/70 (6%)
Query 49 FIASGGSQYLDTTQIPDFICASNAQVKVKE---YFEGLPADIRLEFNNDPMQFAEVVSDP 105
F+ G Q DTT++P C +V E Y + LPA+ DPM F EV+ +
Sbjct 140 FVVGQGMQTYDTTELPLTACMRQIEVVHAEAVRYLKELPANAFDVVYMDPM-FEEVIEES 198
Query 106 RNTDYLRDIG 115
N + LR G
Sbjct 199 NNFEALRLAG 208
> kse:Ksed_09770 NAD-dependent aldehyde dehydrogenase
Length=461
Score = 34.7 bits (78), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 49/102 (48%), Gaps = 12/102 (12%)
Query 48 PFIASGGSQYLDTTQIPDFICASNAQVKVKEYFE-GLPADIRLEFNNDPMQFAEVVSDPR 106
P I G + L QI CA +A++ + + + GLPA + + Q AEV++DPR
Sbjct 148 PNILLGNTILLKHAQI----CAGSARLMEQVFTDAGLPAGVYTDLRISNEQAAEVIADPR 203
Query 107 NTDYLRDIGVLAPLPAEQEGEKHSAQSGDNSKKAPQPSEGSD 148
+R + + +E+ G + Q+G + KK GSD
Sbjct 204 ----VRGVSLTG---SERAGSAVAEQAGKHLKKVVLELGGSD 238
Lambda K H a alpha
0.312 0.131 0.380 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125369324234