bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-33_CDS_annotation_glimmer3.pl_2_3
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
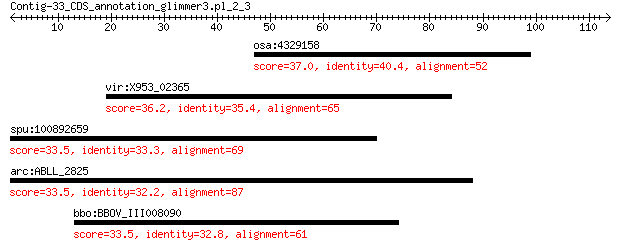
osa:4329158 Os02g0318500 37.0 0.49
vir:X953_02365 hypothetical protein 36.2 0.77
spu:100892659 uncharacterized LOC100892659 33.5 6.3
arc:ABLL_2825 hypothetical protein 33.5 6.7
bbo:BBOV_III008090 17.m07709; hypothetical protein 33.5 8.4
> osa:4329158 Os02g0318500
Length=1315
Score = 37.0 bits (84), Expect = 0.49, Method: Composition-based stats.
Identities = 21/57 (37%), Positives = 28/57 (49%), Gaps = 6/57 (11%)
Query 47 KNQLSLQELVFSELGYSDELF-----DSFYIKPCIKKLKNSYTDKWKDTNYREVHYF 98
KNQ ++EL GY D LF SFYI+ C+ L Y WK+ +Y + Y
Sbjct 1009 KNQELIEELSIPPPGYRDLLFATKYSQSFYIQ-CVANLWKQYKSYWKNPSYNSLRYL 1064
> vir:X953_02365 hypothetical protein
Length=408
Score = 36.2 bits (82), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 41/67 (61%), Gaps = 3/67 (4%)
Query 19 RFDKMKGSLRSKLKAVALFYDYRDYESLKN-QLSLQELVFS-ELGYSDELFDSFYIKPCI 76
+F KG ++ +K + L DYR S + ++ ++ V+S EL Y +L +SFY +P I
Sbjct 36 KFKVEKGDIKD-IKNLKLVGDYRQGMSAEGVAITSEDTVYSSELSYFSQLNNSFYTEPAI 94
Query 77 KKLKNSY 83
++L+N+Y
Sbjct 95 QQLQNNY 101
> spu:100892659 uncharacterized LOC100892659
Length=1860
Score = 33.5 bits (75), Expect = 6.3, Method: Composition-based stats.
Identities = 23/72 (32%), Positives = 37/72 (51%), Gaps = 7/72 (10%)
Query 1 LDWSVPVGKISRFFYRFNRFDKMKGSLRSKLKAVALFYDYRDYES---LKNQLSLQELVF 57
+ W V ++ F Y+F DK + + R L ++ YD Y S LK ++ LQEL
Sbjct 1061 ITWDV---EMDSFLYKFTPRDKPR-TRRGLLSVISSVYDPLGYASPFVLKAKMILQELTR 1116
Query 58 SELGYSDELFDS 69
LG+ D++ D+
Sbjct 1117 MNLGWDDKIPDA 1128
> arc:ABLL_2825 hypothetical protein
Length=454
Score = 33.5 bits (75), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 46/96 (48%), Gaps = 11/96 (11%)
Query 1 LDWSVPVGKISRFFYRFNRFDKMKGSLRSKLKAVALFYD--YRDYESLKNQLSLQELVFS 58
++ S+P I R FDKM L ++KA+ LF + Y D + +K+ + +E +F
Sbjct 47 IERSLPFDVIPRII-DSKEFDKMDKGLSQRIKALNLFLEDLYTDKKIVKDGIIPEEFIFQ 105
Query 59 ELGYSDELFDSFYIKPCIKK-------LKNSYTDKW 87
GY EL + F I+ +K++ TD W
Sbjct 106 AKGYLKEL-EGFSPNKKIRTHINGIDLVKDTVTDDW 140
> bbo:BBOV_III008090 17.m07709; hypothetical protein
Length=1340
Score = 33.5 bits (75), Expect = 8.4, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 36/62 (58%), Gaps = 7/62 (11%)
Query 13 FFYRFNRFDKMKGSLRSKLKAVALFYDYRDYESLKNQL-SLQELVFSELGYSDELFDSFY 71
F Y F+ G+LRS +++ A+F D RD +++ LQ ++ + + DELFDS+
Sbjct 433 FCYTFD------GALRSTVRSAAIFSDQRDDGHIEDAFRKLQVILEDCVSHCDELFDSWD 486
Query 72 IK 73
++
Sbjct 487 LR 488
Lambda K H a alpha
0.323 0.140 0.424 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126991827136