bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-34_CDS_annotation_glimmer3.pl_2_1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
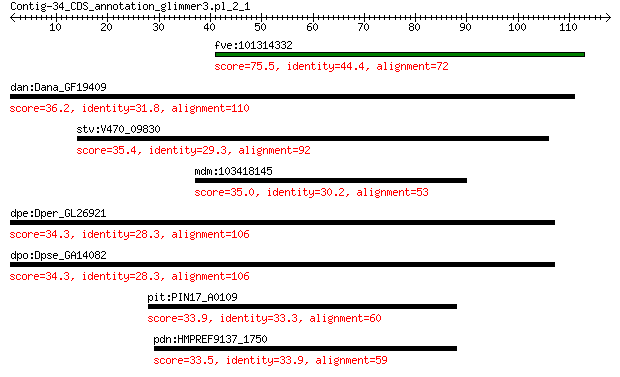
fve:101314332 capsid protein VP1-like 75.5 4e-14
dan:Dana_GF19409 GF19409 gene product from transcript GF19409-RA 36.2 0.73
stv:V470_09830 capsid protein 35.4 1.8
mdm:103418145 protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like 35.0 2.0
dpe:Dper_GL26921 GL26921 gene product from transcript GL26921-RA 34.3 3.2
dpo:Dpse_GA14082 GA14082 gene product from transcript GA14082-RA 34.3 3.2
pit:PIN17_A0109 lpxB; lipid-A-disaccharide synthase (EC:2.4.1.... 33.9 5.4
pdn:HMPREF9137_1750 lpxB; lipid-A-disaccharide synthase (EC:2.... 33.5 7.9
> fve:101314332 capsid protein VP1-like
Length=421
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 32/72 (44%), Positives = 48/72 (67%), Gaps = 2/72 (3%)
Query 41 VLPGDSFNISTSKVVRSQTMLTPIMDNMFLDTYYFFVPNRLVWKHWREFCGENREGAWAP 100
VLPGD+FN++ + R T + P+MDN+ LD+++FFVPNRLVW +W +F GE A
Sbjct 77 VLPGDTFNVNVTMFGRLATPIFPVMDNLHLDSFFFFVPNRLVWNNWVKFMGEQDNP--AD 134
Query 101 TVEYTVPSIAPP 112
++ Y++P P
Sbjct 135 SISYSIPQQVSP 146
> dan:Dana_GF19409 GF19409 gene product from transcript GF19409-RA
Length=254
Score = 36.2 bits (82), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 51/115 (44%), Gaps = 11/115 (10%)
Query 1 VESHFAQLPAAEIQRSTFDRS-HGYKTSFCAGDIIPF---MVDEVLPGDSFNISTSKVVR 56
+E HF ++ EI+R R+ H + G I + M+D+ LPG S I K+V
Sbjct 104 LEQHF-EIYCGEIERFESTRTGHMAISGVTVGIICHYWYKMLDKRLPGRSMRIVAKKIVL 162
Query 57 SQTMLTPIMDNMFLDTYYFFVPNRLVWKHWREFCGENREGAWAP-TVEYTVPSIA 110
Q + +PI + F F L K E E +E AW E+TV +A
Sbjct 163 DQLICSPIYISAF-----FVTLGLLERKDKNEVWAEIKEKAWKLYAAEWTVWPVA 212
> stv:V470_09830 capsid protein
Length=427
Score = 35.4 bits (80), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 42/96 (44%), Gaps = 7/96 (7%)
Query 14 QRSTFDRSHGYKTSFCAGDI---IPFMVDEVLPGDSFNISTSKVVRSQTMLTPIMDNMFL 70
+R D SH F AG I I V+ GDSF + +R + + + +
Sbjct 9 ERMPHDLSH---LGFLAGQIGRLITISTTPVIAGDSFEMDAVGALRLSPLRRGLAIDSTV 65
Query 71 DTYYFFVPNRLVW-KHWREFCGENREGAWAPTVEYT 105
D + F+VP+R V+ + W +F + PTV T
Sbjct 66 DIFTFYVPHRHVYGEQWIKFMKDGVNATPLPTVNTT 101
> mdm:103418145 protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like
Length=853
Score = 35.0 bits (79), Expect = 2.0, Method: Composition-based stats.
Identities = 16/56 (29%), Positives = 30/56 (54%), Gaps = 3/56 (5%)
Query 37 MVDEVLPGDSFNISTSKVV---RSQTMLTPIMDNMFLDTYYFFVPNRLVWKHWREF 89
+ D +PGD ++ + +V+ R+ + P + + L+ Y +P L+W HW EF
Sbjct 643 LADNEIPGDLGSLISLQVLDLSRNNFHILPNLSGVELNGVYKHLPKELIWLHWEEF 698
> dpe:Dper_GL26921 GL26921 gene product from transcript GL26921-RA
Length=239
Score = 34.3 bits (77), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 50/111 (45%), Gaps = 11/111 (10%)
Query 1 VESHFAQLPAAEIQR-STFDRSHGYKTSFCAGDIIPF---MVDEVLPGDSFNISTSKVVR 56
+E H ++ + EI+R + SH + G I F M+D+ +PG S + K+V
Sbjct 89 MEQHL-EIYSGEIERFDSLRTSHMATSGVTVGIICHFWYKMLDKRMPGRSMRVVAKKIVL 147
Query 57 SQTMLTPIMDNMFLDTYYFFVPNRLVWKHWREFCGENREGAWAP-TVEYTV 106
Q + +P+ ++F F L K E E ++ AW E+TV
Sbjct 148 DQLICSPVYISVF-----FVTLGLLEQKDKHEVWDEIKDKAWKLYAAEWTV 193
> dpo:Dpse_GA14082 GA14082 gene product from transcript GA14082-RA
Length=239
Score = 34.3 bits (77), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 50/111 (45%), Gaps = 11/111 (10%)
Query 1 VESHFAQLPAAEIQR-STFDRSHGYKTSFCAGDIIPF---MVDEVLPGDSFNISTSKVVR 56
+E H ++ + EI+R + SH + G I F M+D+ +PG S + K+V
Sbjct 89 MEQHL-EIYSGEIERFDSLRTSHMATSGVTVGIICHFWYKMLDKRMPGRSMRVVAKKIVL 147
Query 57 SQTMLTPIMDNMFLDTYYFFVPNRLVWKHWREFCGENREGAWAP-TVEYTV 106
Q + +P+ ++F F L K E E ++ AW E+TV
Sbjct 148 DQLICSPVYISVF-----FVTLGLLEQKDKHEVWDEIKDKAWKLYAAEWTV 193
> pit:PIN17_A0109 lpxB; lipid-A-disaccharide synthase (EC:2.4.1.182)
Length=380
Score = 33.9 bits (76), Expect = 5.4, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 28/62 (45%), Gaps = 11/62 (18%)
Query 28 FCAGDIIPFMVDEVLPGD--SFNISTSKVVRSQTMLTPIMDNMFLDTYYFFVPNRLVWKH 85
FC DI+ + D V+ D FN+ +K V T + P+ YY+ P WK
Sbjct 75 FCKEDIVKWQPDVVILVDYPGFNLKIAKFVHKNTKI-PV--------YYYISPKIWAWKE 125
Query 86 WR 87
WR
Sbjct 126 WR 127
> pdn:HMPREF9137_1750 lpxB; lipid-A-disaccharide synthase (EC:2.4.1.182)
Length=386
Score = 33.5 bits (75), Expect = 7.9, Method: Composition-based stats.
Identities = 20/61 (33%), Positives = 29/61 (48%), Gaps = 11/61 (18%)
Query 29 CAGDIIPFMVDEVLPGD--SFNISTSKVVRSQTMLTPIMDNMFLDTYYFFVPNRLVWKHW 86
C DI+ + D V+ D FN+S +K V+ T + P+ YY+ P WK W
Sbjct 76 CKADIVRWKPDVVILVDYPGFNLSIAKFVKKNTDI-PV--------YYYISPKIWAWKEW 126
Query 87 R 87
R
Sbjct 127 R 127
Lambda K H a alpha
0.324 0.138 0.463 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125169047616