bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-39_CDS_annotation_glimmer3.pl_2_1
Length=322
Score E
Sequences producing significant alignments: (Bits) Value
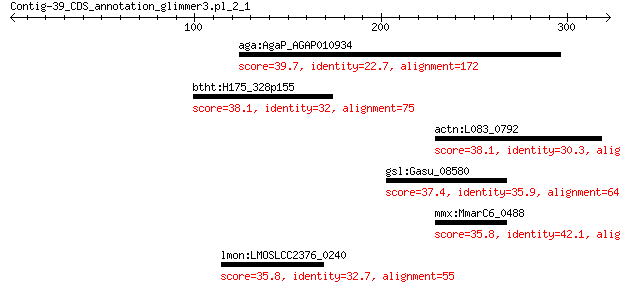
aga:AgaP_AGAP010934 AGAP010934-PA 39.7 1.0
btht:H175_328p155 N-hydroxyarylamine O-acetyltransferase 38.1 1.9
actn:L083_0792 endo-1,4-beta-xylanase 38.1 1.9
gsl:Gasu_08580 cation transport protein ChaC 37.4 2.4
mmx:MmarC6_0488 hypothetical protein 35.8 5.1
lmon:LMOSLCC2376_0240 phosphoglycerate mutase family protein (... 35.8 7.3
> aga:AgaP_AGAP010934 AGAP010934-PA
Length=1919
Score = 39.7 bits (91), Expect = 1.0, Method: Composition-based stats.
Identities = 39/181 (22%), Positives = 76/181 (42%), Gaps = 24/181 (13%)
Query 124 WQLTKTLFENWIDGYR--FYGSYVNEKTINYVSKYMTKKDEDNPDYIGIVLCSKGLGANY 181
W+LT T + R ++ ++V TI ++ K + E NP I +L + + +
Sbjct 703 WKLTMTWANKFYREVRSSWHSNHVESVTIEWMEKRLL---EYNPSVIAGILKNGSISMDC 759
Query 182 AKRMAYKHEWNK-------EKTNITYKAKNGADLPLPRYYKTQLYTEDQRQLLWLYAEDE 234
A+ Y K E I +A + L YY + +++ ++L + ++
Sbjct 760 AEEFDYVEHTRKLLEQLGLEVDTIGEEALQQLNKRLKPYYNNMFFVDNKWKVLEAFCKER 819
Query 235 GVKWVKGFKVQGANGANKDYYETLVKQKNEEGAPLHGDVEEEIIRKKAINRMYKLQSLTQ 294
+ W +K+ TLV++ EE L+ + EE+ + N ++ L LT+
Sbjct 820 KLVW------------SKELTRTLVRKDQEELQQLYDERREELRQLLKANELHSLDGLTR 867
Query 295 R 295
R
Sbjct 868 R 868
> btht:H175_328p155 N-hydroxyarylamine O-acetyltransferase
Length=268
Score = 38.1 bits (87), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 37/75 (49%), Gaps = 7/75 (9%)
Query 99 TEKGHTNTRRIHIHGLYYATHGETKWQLTKTLFENWIDGYRFYGSYVNEKTINYVSKYMT 158
TEKG H+ + +GET L L +NW GY FY + +NEK +N + M
Sbjct 157 TEKG------THVFEMKKGKNGETSHFLDSDLTDNWSIGYAFYLNVINEKKVNAIQN-MV 209
Query 159 KKDEDNPDYIGIVLC 173
+ ++P G ++C
Sbjct 210 IEHPESPVNKGNIIC 224
> actn:L083_0792 endo-1,4-beta-xylanase
Length=363
Score = 38.1 bits (87), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 38/89 (43%), Gaps = 10/89 (11%)
Query 229 LYAEDEGVKWVKGFKVQGANGANKDYYETLVKQKNEEGAPLHGDVEEEIIRKKAINRMYK 288
L+ D GV+W GA D YE L +Q +G PLHG + + + +
Sbjct 208 LFLNDYGVEW---------PGAKVDAYEALARQLLADGVPLHGFAAQAHLSMQ-YGAPDQ 257
Query 289 LQSLTQRKQNQKRTIKREEKDLFYSLKDG 317
LQS+ QR N E D+ +L G
Sbjct 258 LQSVLQRFDNLGLQTAITELDVRMTLPAG 286
> gsl:Gasu_08580 cation transport protein ChaC
Length=201
Score = 37.4 bits (85), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/64 (36%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 203 KNGADLPLPRYYKTQLYTEDQRQLLWLYAEDEGVKWVKGFKVQGANGANKDYYETLVKQK 262
K A+LP+ R + +E RQ WL DE V FK +G +G+N++Y+E LV
Sbjct 118 KEDAELPVVRDALIYIASETNRQ--WLGPSDEKVMIEHIFKSRGPSGSNREYFEKLVDSM 175
Query 263 NEEG 266
G
Sbjct 176 RSFG 179
> mmx:MmarC6_0488 hypothetical protein
Length=148
Score = 35.8 bits (81), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 26/38 (68%), Gaps = 2/38 (5%)
Query 229 LYAEDEGVKWVKGFKVQGANGANKDYYETLVKQKNEEG 266
L+ EDE +++V GF VQ +G +D +E +V++ EEG
Sbjct 99 LFTEDEELQYVLGFAVQDVSG--RDRFEIIVEKPEEEG 134
> lmon:LMOSLCC2376_0240 phosphoglycerate mutase family protein
(EC:5.4.2.-)
Length=211
Score = 35.8 bits (81), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 18/63 (29%), Positives = 30/63 (48%), Gaps = 8/63 (13%)
Query 114 LYYATHGETKWQLT--------KTLFENWIDGYRFYGSYVNEKTINYVSKYMTKKDEDNP 165
+Y+ HG+T+W +T L E IDG + G + + I+ V +K+ +D
Sbjct 8 IYFVRHGKTEWNMTGQMQGWGDSPLVEEGIDGAKAVGEVLRDTPIDAVYTSTSKRTQDTA 67
Query 166 DYI 168
YI
Sbjct 68 AYI 70
Lambda K H a alpha
0.318 0.136 0.422 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 575086215924