bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
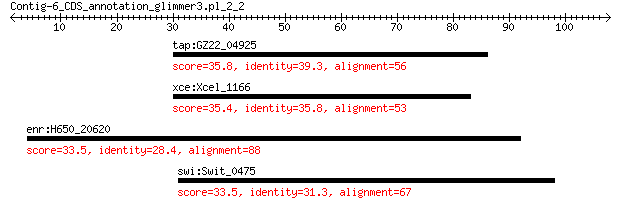
tap:GZ22_04925 penicillin-binding protein 35.8 0.94
xce:Xcel_1166 amidohydrolase 35.4 1.1
enr:H650_20620 nucleotide-binding protein 33.5 4.0
swi:Swit_0475 5-oxoprolinase (EC:3.5.2.9) 33.5 5.3
> tap:GZ22_04925 penicillin-binding protein
Length=727
Score = 35.8 bits (81), Expect = 0.94, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 5/61 (8%)
Query 30 GLTPQQVAE----MAKRGIPVSPMNVNFI-DVNGDASWNIEPQFRRDMDMATAWEMEKAS 84
GLTP ++AE A G + ++N I D NG+A++ EPQ TAW+M +
Sbjct 467 GLTPLKIAEGFRTFAHEGQWIESQSINAIYDRNGEAAFKAEPQTEDVFSEQTAWDMTRML 526
Query 85 Q 85
Q
Sbjct 527 Q 527
> xce:Xcel_1166 amidohydrolase
Length=373
Score = 35.4 bits (80), Expect = 1.1, Method: Composition-based stats.
Identities = 19/55 (35%), Positives = 27/55 (49%), Gaps = 2/55 (4%)
Query 30 GLTPQQVAEMAKRGIPVSP--MNVNFIDVNGDASWNIEPQFRRDMDMATAWEMEK 82
G+TP Q E+A RG+PV+P + V V D + P F R + A E+
Sbjct 216 GVTPDQAVELAARGVPVTPTLLQVERFGVIADQAQERYPAFARRLRAMHARRYEQ 270
> enr:H650_20620 nucleotide-binding protein
Length=163
Score = 33.5 bits (75), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 51/96 (53%), Gaps = 9/96 (9%)
Query 4 NTKPDYKCVECNFDVQKDFERTKPNLGLTPQQV--------AEMAKRGIPVSPMNVNFID 55
T+ D++ VE F++ ++ ++T L + QV A++ KRGI + ++V
Sbjct 27 GTRFDFRNVEATFELNEE-KQTIKVLSESDFQVNQLLDILRAKLLKRGIEGASLDVPEEF 85
Query 56 VNGDASWNIEPQFRRDMDMATAWEMEKASQRKALQV 91
V+ +W +E + ++ +D ATA ++ K + L+V
Sbjct 86 VHSGKTWFVEAKLKQGIDAATAKKIIKLIKDSKLKV 121
> swi:Swit_0475 5-oxoprolinase (EC:3.5.2.9)
Length=680
Score = 33.5 bits (75), Expect = 5.3, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 39/74 (53%), Gaps = 7/74 (9%)
Query 31 LTPQQVAEMAKRGIPVSPMNVNFI----DVNGDASWNIEPQFRRDM-DMATAWEMEK--A 83
L PQ+ + RGI +S +N++F+ + DASW +M D ATAW + + A
Sbjct 468 LVPQEPGTLCARGILLSDINMDFVRSELSLAADASWRRVCAVLGEMRDEATAWLVREGVA 527
Query 84 SQRKALQVLRQKKF 97
+ + L+++ ++
Sbjct 528 EEDRQLRIMVDARY 541
Lambda K H a alpha
0.317 0.131 0.398 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125230604613