bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_6
Length=59
Score E
Sequences producing significant alignments: (Bits) Value
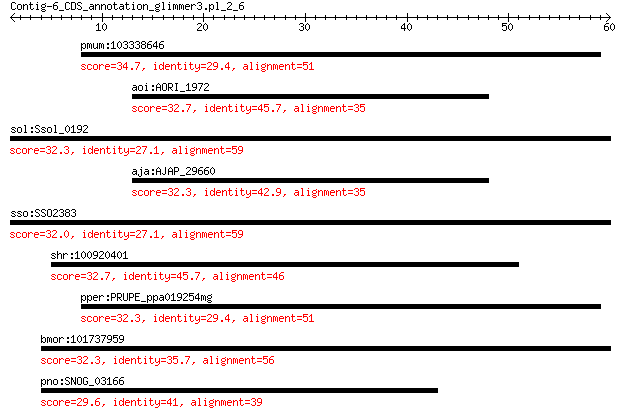
pmum:103338646 uncharacterized LOC103338646 34.7 0.84
aoi:AORI_1972 carotenoid cleavage dioxygenase 32.7 4.5
sol:Ssol_0192 NMD3 family protein 32.3 4.7
aja:AJAP_29660 Carotenoid cleavage oxygenase (EC:1.13.11.-) 32.3 4.8
sso:SSO2383 hypothetical protein 32.0 5.3
shr:100920401 ankyrin repeat domain-containing protein 26-like 32.7 5.3
pper:PRUPE_ppa019254mg hypothetical protein 32.3 5.4
bmor:101737959 sulfhydryl oxidase 1-like 32.3 6.0
pno:SNOG_03166 hypothetical protein 29.6 8.1
> pmum:103338646 uncharacterized LOC103338646
Length=331
Score = 34.7 bits (78), Expect = 0.84, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 30/51 (59%), Gaps = 0/51 (0%)
Query 8 FEIIWRDPKTGQFKTSEYRKKNQMSIVDARSYARRLVNIQNVLSVRFYKEM 58
FE++W++P++ +K E +K + I+ + +R LV + L V F K++
Sbjct 189 FEVLWKEPRSIIYKVGEIERKIEFLILRMKFNSRCLVEVPEYLGVNFEKQI 239
> aoi:AORI_1972 carotenoid cleavage dioxygenase
Length=480
Score = 32.7 bits (73), Expect = 4.5, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (59%), Gaps = 4/39 (10%)
Query 13 RDPKTGQFKTSEY----RKKNQMSIVDARSYARRLVNIQ 47
RDP+TG+ Y K Q S++DA+ ARR V+I+
Sbjct 155 RDPETGELHAVSYFFGWGNKVQYSVIDAQGRARRKVDIE 193
> sol:Ssol_0192 NMD3 family protein
Length=237
Score = 32.3 bits (72), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 32/59 (54%), Gaps = 10/59 (17%)
Query 1 MNKYTFIFEIIWRDPKTGQFKTSEYRKKNQMSIVDARSYARRLVNIQNVLSVRFYKEMC 59
++++ F + IW+DP F T E+R K V +S+++ + V+S+ +E+C
Sbjct 85 VDEFAFDIKSIWKDPSGNSFATIEFRGK-----VRGKSFSQ-----EAVISLEMERELC 133
> aja:AJAP_29660 Carotenoid cleavage oxygenase (EC:1.13.11.-)
Length=478
Score = 32.3 bits (72), Expect = 4.8, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 23/39 (59%), Gaps = 4/39 (10%)
Query 13 RDPKTGQFKTSEY----RKKNQMSIVDARSYARRLVNIQ 47
RDP+TG+ Y K Q S++DA+ ARR V+++
Sbjct 152 RDPETGELHAVSYFFGSGNKVQYSVIDAQGRARRTVDVE 190
> sso:SSO2383 hypothetical protein
Length=240
Score = 32.0 bits (71), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 32/59 (54%), Gaps = 10/59 (17%)
Query 1 MNKYTFIFEIIWRDPKTGQFKTSEYRKKNQMSIVDARSYARRLVNIQNVLSVRFYKEMC 59
++++ F + IW+DP F T E+R K V +S+++ + V+S+ +E+C
Sbjct 88 VDEFAFDIKSIWKDPSGNSFATIEFRGK-----VRGKSFSQ-----EAVISLEMERELC 136
> shr:100920401 ankyrin repeat domain-containing protein 26-like
Length=1712
Score = 32.7 bits (73), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 27/55 (49%), Gaps = 9/55 (16%)
Query 5 TFIFEIIWRDPKTGQFKTSEYRKKNQMS-------IVDARSYARRLVNIQ--NVL 50
T I E + R+ K Q+K +E + KNQM IV S RL IQ N+L
Sbjct 1175 TVIIESVQRELKQAQYKATELKHKNQMDNEKLDRYIVKQESIQERLAQIQSENIL 1229
> pper:PRUPE_ppa019254mg hypothetical protein
Length=455
Score = 32.3 bits (72), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 29/51 (57%), Gaps = 0/51 (0%)
Query 8 FEIIWRDPKTGQFKTSEYRKKNQMSIVDARSYARRLVNIQNVLSVRFYKEM 58
FE++W++P++ +K E +K + I + +R LV + L V F K++
Sbjct 313 FEVLWKEPRSIIYKVGEIERKIEFLIRRMKFNSRCLVEVPEYLGVNFEKQI 363
> bmor:101737959 sulfhydryl oxidase 1-like
Length=571
Score = 32.3 bits (72), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 30/64 (47%), Gaps = 8/64 (13%)
Query 4 YTFIFEIIWRDPKTGQFKTSE--------YRKKNQMSIVDARSYARRLVNIQNVLSVRFY 55
YT IFEI TG + YRK +Q+ I+ +++ ++L N L V+FY
Sbjct 5 YTSIFEIFLIALVTGAVVPTTDDVDEQGLYRKSDQVEILTNKNFEKKLYGQNNALLVQFY 64
Query 56 KEMC 59
C
Sbjct 65 NSYC 68
> pno:SNOG_03166 hypothetical protein
Length=45
Score = 29.6 bits (65), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 1/40 (3%)
Query 4 YTFIFEIIWRDPKTGQFKTSEYRKKNQMSIVDA-RSYARR 42
Y II +DPK G+ K Y K+N+ SI+ R+Y R
Sbjct 6 YISHHPIIIQDPKLGRLKVHSYSKRNKKSIMHQLRNYKGR 45
Lambda K H a alpha
0.326 0.136 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 125989489341