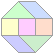
 Emax_3360 (135 letters)
Emax_3360 (135 letters)
5 hits
 135 letters
135 letters
cel 



 63.0 - CE05682 (418) KOG0194
63.0 - CE05682 (418) KOG0194
ath 



 63.0 - At5g45230_1 (1191)
63.0 - At5g45230_1 (1191)
ath 



 61.0 - At5g04810 (949) KOG4197
61.0 - At5g04810 (949) KOG4197
hsa 



 59.0 - Hs4506303 (802) KOG4228
59.0 - Hs4506303 (802) KOG4228
hsa 



 59.0 - Hs18450369 (793) KOG4228
59.0 - Hs18450369 (793) KOG4228
BLASTP 2.2.18 Mar-02-2008
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query: Emax_3360 (135 letters)
Database: /usr/local/genome/databases/kog/kyva
47500486 sequences; 112920 total letters
>gnl|BL_ORD_ID|46730 CE05682
Length = 418
Score: 63.0 Expect: 2.63734
Identities 16 (34%) Positives: 23 (48%) Gaps: 1/47 (2%)
Query: 82 SKVCCGELSVTFPETVPPSWSNEDADLARLLLSLTRQAMHQLQHHQL 128
+K+ C + +TFPET PP+ S L R M + HH+L
Sbjct: 355 AKIFCDDYRMTFPETTPPTISEIALKQCWAKLPSDRATMKTV-HHKL 400
>gnl|BL_ORD_ID|1219 At5g45230_1
Length = 1191
Score: 63.0 Expect: 3.22205
Identities 18 (31%) Positives: 24 (41%) Gaps: 1/58 (1%)
Query: 45 LGKQVAVTIQRKQQLRLVVEFSLPFEEAKQDGEHSHCSKVCCGELSVTFP-ETVPPSW 101
+G + I+ +QQ L E SL F+ E C + CG V P E SW
Sbjct: 1058 IGYTTLLNIKNRQQFPLATEISLRFQVTNGTSEVEKCKVIKCGFSLVYEPNEADSTSW 1115
>gnl|BL_ORD_ID|37153 At5g04810
Length = 949
Score: 61.0 Expect: 4.72956
Identities 14 (41%) Positives: 17 (50%) Gaps: 6/34 (17%)
Query: 76 GEHSHCSKVCC------GELSVTFPETVPPSWSN 103
G H SK C GEL+ T +T PP WS+
Sbjct: 873 GTAVHWSKCLCKIEASGGELTETLQKTFPPDWSS 906
>gnl|BL_ORD_ID|94461 Hs4506303
Length = 802
Score: 59.0 Expect: 9.29685
Identities 12 (31%) Positives: 22 (57%) Gaps: 0/38 (0%)
Query: 61 LVVEFSLPFEEAKQDGEHSHCSKVCCGELSVTFPETVP 98
++V + L F++ KQ G HS+ ++ G P++VP
Sbjct: 169 IIVLYMLRFKKYKQAGSHSNSFRLSNGRTEDVEPQSVP 206
>gnl|BL_ORD_ID|73015 Hs18450369
Length = 793
Score: 59.0 Expect: 9.37474
Identities 12 (31%) Positives: 22 (57%) Gaps: 0/38 (0%)
Query: 61 LVVEFSLPFEEAKQDGEHSHCSKVCCGELSVTFPETVP 98
++V + L F++ KQ G HS+ ++ G P++VP
Sbjct: 160 IIVLYMLRFKKYKQAGSHSNSFRLSNGRTEDVEPQSVP 197
Database: /usr/local/genome/databases/kog/kyva
Number of letters in database: 47500486
Number of sequences in database: 112920
Matrix: BLOSUM62
Gap penalties: Existence: 11 Extension: 1
Number of sequences: 112920
Length of database: 47500486
Effecttive HSP length: 0
Effecttive length of query: 135
Effective search space: 0.0
Effective search space used: 0.0
T: 11
A: 40
X1: 16 (7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (20.4 bits)
S2: 73 (32.7 bits)
 Emax_3360 (135 letters)
Emax_3360 (135 letters) Emax_3360 (135 letters)
Emax_3360 (135 letters) 135 letters
135 letters



 63.0 - CE05682 (418) KOG0194
63.0 - CE05682 (418) KOG0194



 63.0 - At5g45230_1 (1191)
63.0 - At5g45230_1 (1191)



 61.0 - At5g04810 (949) KOG4197
61.0 - At5g04810 (949) KOG4197



 59.0 - Hs4506303 (802) KOG4228
59.0 - Hs4506303 (802) KOG4228



 59.0 - Hs18450369 (793) KOG4228
59.0 - Hs18450369 (793) KOG4228