bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0005_orf2
Length=230
Score E
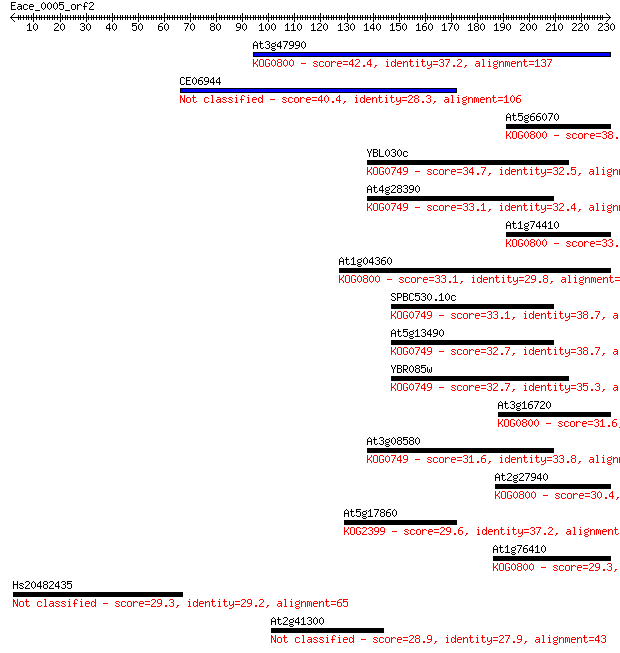
Sequences producing significant alignments: (Bits) Value
At3g47990 42.4 8e-04
CE06944 40.4 0.003
At5g66070 38.9 0.008
YBL030c 34.7 0.19
At4g28390 33.1 0.43
At1g74410 33.1 0.44
At1g04360 33.1 0.51
SPBC530.10c 33.1 0.54
At5g13490 32.7 0.56
YBR085w 32.7 0.57
At3g16720 31.6 1.4
At3g08580 31.6 1.5
At2g27940 30.4 3.5
At5g17860 29.6 5.0
At1g76410 29.3 6.4
Hs20482435 29.3 6.5
At2g41300 28.9 9.3
> At3g47990
Length=292
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 51/175 (29%), Positives = 66/175 (37%), Gaps = 46/175 (26%)
Query 94 FPCFFGWTVVGTFWFLEVQEETPFCLPRESHPWFFTFWLALCYVWILIYIVFIVMAAVFE 153
+P + WTV+GT WF + + CLP E W F WL Y +L I FI V +
Sbjct 17 YPFLWAWTVIGTQWFTKSKT----CLPEEGQKWGFLIWLMFSYCGLLC-IAFI---CVGK 68
Query 154 YRARRMEADLRLLQN---------EDMIR--RWG------RVRVFSE------YGIYIF- 189
+ RR LR Q DMIR W +R S+ G+Y+
Sbjct 69 WLTRRQVHLLRAQQGIPISEFGILVDMIRVPDWAFEAAGQEMRGISQDAATYHPGLYLTP 128
Query 190 --------------RRGLTTDQIRRLPCQKLNYDPVEMMPCSICLEEFAQDEYVR 230
R I+ LP +L P + C ICLEEF VR
Sbjct 129 AQASLVIYSQQAKQRTEAVEALIQELPKFRLKAVPDDCGECLICLEEFHIGHEVR 183
> CE06944
Length=263
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 53/109 (48%), Gaps = 3/109 (2%)
Query 66 LSEDGEDFMLYRQRGPPFWVNV-LILGILFPCFFGWTVVGTFWFLEVQEETPFCLPRES- 123
LS D +LY + F V V ++ L P +G VG + V+E P C P +
Sbjct 40 LSVDLLITILYPIKSRNFHVPVYFVILFLLPVAYGAIFVGFGFMYLVEEALPMCNPPSAL 99
Query 124 HPWFFTFWLALCYVWILIYIVFIVMA-AVFEYRARRMEADLRLLQNEDM 171
HP ++W + + ++ ++F V A A+ ++ RR DLR ++ + +
Sbjct 100 HPIVRSYWYYVMTAFSVLTVIFYVSAFALIYFKGRRENTDLRFIERKAL 148
> At5g66070
Length=221
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 191 RGLTTDQIRRLPCQKLNYDPVEMMPCSICLEEFAQDEYVR 230
+GLT D + R+P ++ E++ CS+CL++F E VR
Sbjct 151 KGLTGDSLNRIPKVRITDTSPEIVSCSVCLQDFQVGETVR 190
> YBL030c
Length=318
Score = 34.7 bits (78), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 45/92 (48%), Gaps = 15/92 (16%)
Query 138 WILIYIVFIVMAAVFEYRARRMEADLRLLQNEDMIRRWGRV------------RVFSEYG 185
+++ +++ V AAV + A +E L+QN+D + + G + R ++ G
Sbjct 23 FLIDFLMGGVSAAVAKTAASPIERVKLLIQNQDEMLKQGTLDRKYAGILDCFKRTATQEG 82
Query 186 IYIFRRGLTTDQIRRLPCQKLNY---DPVEMM 214
+ F RG T + IR P Q LN+ D ++ M
Sbjct 83 VISFWRGNTANVIRYFPTQALNFAFKDKIKAM 114
> At4g28390
Length=379
Score = 33.1 bits (74), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 40/83 (48%), Gaps = 12/83 (14%)
Query 138 WILIYIVFIVMAAVFEYRARRMEADLRLLQNEDMIRRWGRV------------RVFSEYG 185
+++ +++ V AAV + A +E L+QN+D + + GR+ R + G
Sbjct 79 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGISDCFARTVKDEG 138
Query 186 IYIFRRGLTTDQIRRLPCQKLNY 208
+ RG T + IR P Q LN+
Sbjct 139 MLALWRGNTANVIRYFPTQALNF 161
> At1g74410
Length=106
Score = 33.1 bits (74), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 26/42 (61%), Gaps = 2/42 (4%)
Query 191 RGLTTDQIRRLPCQKLNYDPV--EMMPCSICLEEFAQDEYVR 230
RGL+ D +R+LPC ++ + V ++ C+ICL++ E R
Sbjct 32 RGLSGDSLRKLPCYIMSSEMVRRQVTHCTICLQDIKTGEITR 73
> At1g04360
Length=381
Score = 33.1 bits (74), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 41/113 (36%), Gaps = 34/113 (30%)
Query 127 FFTFWLALCYVWILIYIVFIVMAAVFEYRARRMEADLRLLQNEDMIRRWGRVRVFSEYGI 186
++ F + C W I I +R RR +D QN MI Y
Sbjct 61 YYIFVIKCCLNWHQIDI----------FRRRRRSSD----QNPLMI-----------YSP 95
Query 187 YIFRRGLTTDQIRRLPCQKLNYDPV---------EMMPCSICLEEFAQDEYVR 230
+ RGL IR +P K V CS+CL EF +DE +R
Sbjct 96 HEVNRGLDESAIRAIPVFKFKKRDVVAGEEDQSKNSQECSVCLNEFQEDEKLR 148
> SPBC530.10c
Length=322
Score = 33.1 bits (74), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 35/74 (47%), Gaps = 12/74 (16%)
Query 147 VMAAVFEYRARRMEADLRLLQNEDMIRRWGRV------------RVFSEYGIYIFRRGLT 194
V AAV + A +E L+QN+D + R GR+ R +E G+ RG T
Sbjct 36 VSAAVSKTAAAPIERVKLLIQNQDEMIRAGRLSHRYKGIGECFKRTAAEEGVISLWRGNT 95
Query 195 TDQIRRLPCQKLNY 208
+ +R P Q LN+
Sbjct 96 ANVLRYFPTQALNF 109
> At5g13490
Length=385
Score = 32.7 bits (73), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 34/74 (45%), Gaps = 12/74 (16%)
Query 147 VMAAVFEYRARRMEADLRLLQNEDMIRRWGRV------------RVFSEYGIYIFRRGLT 194
V AAV + A +E L+QN+D + + GR+ R + GI RG T
Sbjct 93 VSAAVSKTAAAPIERVKLLIQNQDEMLKAGRLTEPYKGIRDCFGRTIRDEGIGSLWRGNT 152
Query 195 TDQIRRLPCQKLNY 208
+ IR P Q LN+
Sbjct 153 ANVIRYFPTQALNF 166
> YBR085w
Length=307
Score = 32.7 bits (73), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 39/83 (46%), Gaps = 15/83 (18%)
Query 147 VMAAVFEYRARRMEADLRLLQNEDMIRRWGRV------------RVFSEYGIYIFRRGLT 194
V AA+ + A +E L+QN+D + + G + R + G+ F RG T
Sbjct 21 VSAAIAKTAASPIERVKILIQNQDEMIKQGTLDKKYSGIVDCFKRTAKQEGLISFWRGNT 80
Query 195 TDQIRRLPCQKLNY---DPVEMM 214
+ IR P Q LN+ D +++M
Sbjct 81 ANVIRYFPTQALNFAFKDKIKLM 103
> At3g16720
Length=304
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 7/47 (14%)
Query 188 IFRRGLTTDQIRRLP----CQKLNYDPVEMMPCSICLEEFAQDEYVR 230
+ RGL + I+ LP + + DP+E C++CL EF + E R
Sbjct 90 VASRGLDPNVIKSLPVFTFSDETHKDPIE---CAVCLSEFEESETGR 133
> At3g08580
Length=381
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 12/83 (14%)
Query 138 WILIYIVFIVMAAVFEYRARRMEADLRLLQNEDMIRRWGRV------------RVFSEYG 185
+ L +++ V AAV + A +E L+QN+D + + GR+ R + G
Sbjct 80 FALDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGIGDCFGRTIKDEG 139
Query 186 IYIFRRGLTTDQIRRLPCQKLNY 208
RG T + IR P Q LN+
Sbjct 140 FGSLWRGNTANVIRYFPTQALNF 162
> At2g27940
Length=237
Score = 30.4 bits (67), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 13/51 (25%)
Query 187 YIFRRGLTTDQIRRLP-------CQKLNYDPVEMMPCSICLEEFAQDEYVR 230
Y FRRGL + +R LP ++ N D C ICL +F + E V+
Sbjct 110 YSFRRGLDSQAVRSLPVYRYTKAAKQRNED------CVICLSDFEEGETVK 154
> At5g17860
Length=570
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 129 TFWLALCYVWI-LIYIVFIVMAAVFEYRARRMEADLRLLQNEDM 171
T W+ALCY+ I L+Y+ F+ ++ F+ + R + LR ED+
Sbjct 231 TIWVALCYLSIYLLYVGFLSVSHFFDRKKRMSDQILR--SREDL 272
> At1g76410
Length=185
Score = 29.3 bits (64), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 7/50 (14%)
Query 186 IYIFRRGLTTDQIRRLPCQKLNYDP-----VEMMPCSICLEEFAQDEYVR 230
+ +GL +R LP KL Y P +++ C+ICL EFA + +R
Sbjct 71 VAAANKGLKKKVLRSLP--KLTYSPDSPPAEKLVECAICLTEFAAGDELR 118
> Hs20482435
Length=311
Score = 29.3 bits (64), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 34/69 (49%), Gaps = 5/69 (7%)
Query 2 KKKMVVSGFTLNDAILLFAFLYSSSDVF----VEWDAFAACAKPVHYWLLGSYGALIAFR 57
KK +S + I LF L +D+F + +D + A +KP+HY + S G A
Sbjct 87 SKKKTISYTSCMTQIFLFHLL-GGADIFSLSVMAFDCYMAISKPLHYVTIMSRGQCTALI 145
Query 58 LSHYLGQYL 66
+ ++G ++
Sbjct 146 SASWMGGFV 154
> At2g41300
Length=395
Score = 28.9 bits (63), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 101 TVVGTFWFLEVQEETPFCLPRESHPWFFTFWLALCYVWILIYI 143
T G FW ++TPF HPW F++ + +L+++
Sbjct 292 TETGDFWVALHSKKTPFSRLSMIHPWVGKFFIKTLKMELLVFL 334
Lambda K H
0.333 0.146 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4455485750
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40