bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0014_orf1
Length=207
Score E
Sequences producing significant alignments: (Bits) Value
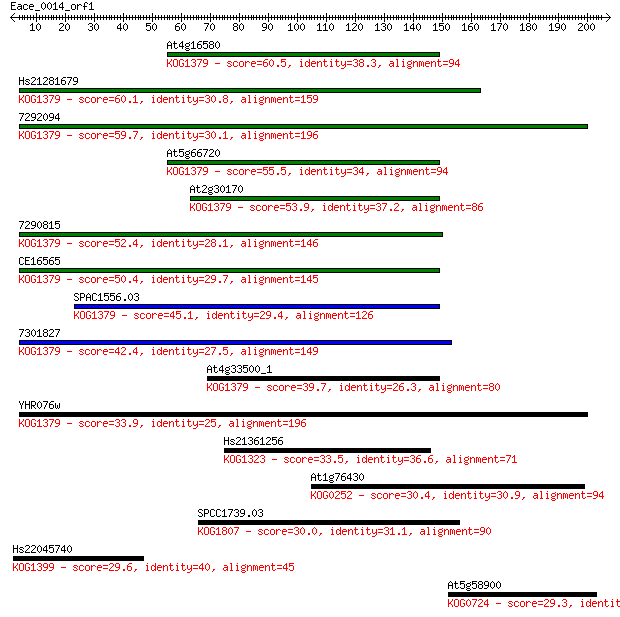
At4g16580 60.5 2e-09
Hs21281679 60.1 3e-09
7292094 59.7 4e-09
At5g66720 55.5 8e-08
At2g30170 53.9 2e-07
7290815 52.4 7e-07
CE16565 50.4 2e-06
SPAC1556.03 45.1 1e-04
7301827 42.4 7e-04
At4g33500_1 39.7 0.004
YHR076w 33.9 0.21
Hs21361256 33.5 0.31
At1g76430 30.4 2.5
SPCC1739.03 30.0 3.2
Hs22045740 29.6 3.9
At5g58900 29.3 6.5
> At4g16580
Length=335
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 53/94 (56%), Gaps = 12/94 (12%)
Query 55 GKIGDEPSAIQSTSVPVQDGDLIILGTDGLFDNVFDFEVLALTGLAVSPHEAQTLLGDAS 114
G+ GD PS+ Q +V V GD+II GTDGLFDN+++ E+ A+ AV +
Sbjct 224 GRNGDLPSSGQVFTVAVAPGDVIIAGTDGLFDNLYNNEITAIVVHAVRAN---------- 273
Query 115 LATPPEDVARALALAAYWRSLDRDAQTPFSKEAR 148
P+ A+ +A A R+ D++ QTPFS A+
Sbjct 274 --IDPQVTAQKIAALARQRAQDKNRQTPFSTAAQ 305
> Hs21281679
Length=304
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 49/160 (30%), Positives = 74/160 (46%), Gaps = 39/160 (24%)
Query 4 LVKKVKEMQHGFNVPFQFAHLPAPEEWDGLLQKGLTRLVGIAKQEYGQGNGGKIGDEPSA 63
+V + E QH FN PFQ + AP E +G++ + D P A
Sbjct 169 VVHRSDEQQHYFNTPFQLS--IAPPEAEGVV----------------------LSDSPDA 204
Query 64 IQSTSVPVQDGDLIILGTDGLFDNVFDFEVL-ALTGLAVSPHEAQTLLGDASLATPPEDV 122
STS VQ GD+I+ TDGLFDN+ D+ +L L L S +E+ +
Sbjct 205 ADSTSFDVQLGDIILTATDGLFDNMPDYMILQELKKLKNSNYES------------IQQT 252
Query 123 ARALALAAYWRSLDRDAQTPFSKEARRCHNKQSLPVQTPD 162
AR++A A+ + D + +PF++ A C N ++ PD
Sbjct 253 ARSIAEQAHELAYDPNYMSPFAQFA--CDNGLNVRGGKPD 290
> 7292094
Length=321
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 59/200 (29%), Positives = 81/200 (40%), Gaps = 72/200 (36%)
Query 4 LVKKVKEMQHGFNVPFQFAHLPAPEEWDGLLQKGLTRLVGIAKQEYGQGNGGKI-GDEPS 62
+V K +E QH FN PFQ + LP P G+G + D P
Sbjct 188 VVHKSEEQQHYFNTPFQLS-LPPP------------------------GHGPNVLSDSPE 222
Query 63 AIQSTSVPVQDGDLIILGTDGLFDNV---FDFEVLALTGLAVSPHEAQTLLGDASLATPP 119
+ + S PV+DGD+I++ TDG+FDNV +VL+ P + Q
Sbjct 223 SADTMSFPVRDGDVILIATDGVFDNVPEDLMLQVLSEVEGERDPVKLQM----------- 271
Query 120 EDVARALALAAYWRSLDRDAQTPFSKEARRCHNKQSLPVQTPDGSSEQGAPQASPVHAMF 179
A +LAL A SL+ + +PF+ ARR +N Q+
Sbjct 272 --TANSLALMARTLSLNSEFLSPFALSARR-NNIQA------------------------ 304
Query 180 LAGLLSGGKEDDITVAAAWV 199
GGK DDITV A V
Sbjct 305 -----RGGKPDDITVVLATV 319
> At5g66720
Length=414
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 53/94 (56%), Gaps = 12/94 (12%)
Query 55 GKIGDEPSAIQSTSVPVQDGDLIILGTDGLFDNVFDFEVLALTGLAVSPHEAQTLLGDAS 114
G D PS+ Q ++ VQ GD+I+ GTDG++DN+++ E+ TG+ VS A
Sbjct 309 GNSADVPSSGQVFTIDVQSGDVIVAGTDGVYDNLYNEEI---TGVVVSSVRA-------- 357
Query 115 LATPPEDVARALALAAYWRSLDRDAQTPFSKEAR 148
P+ A+ +A A R++D+ Q+PF+ A+
Sbjct 358 -GLDPKGTAQKIAELARQRAVDKKRQSPFATAAQ 390
> At2g30170
Length=283
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 49/86 (56%), Gaps = 5/86 (5%)
Query 63 AIQSTSVPVQDGDLIILGTDGLFDNVFDFEVLALTGLAVSPHEAQTLLGDASLATPPEDV 122
A Q + V VQ GD+I++G+DGLFDNVFD E++++ E+ + + T
Sbjct 196 ASQFSIVEVQKGDVIVMGSDGLFDNVFDHEIVSIVTKHTDVAESLSYMFFGITIT----- 250
Query 123 ARALALAAYWRSLDRDAQTPFSKEAR 148
AR LA A S D + ++P++ EAR
Sbjct 251 ARLLAEVASSHSRDTEFESPYALEAR 276
> 7290815
Length=374
Score = 52.4 bits (124), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 41/149 (27%), Positives = 60/149 (40%), Gaps = 40/149 (26%)
Query 4 LVKKVKEMQHGFNVPFQFAHLPAPEEWDGLLQKGLTRLVGIAKQEYGQGNGGKIGDEPSA 63
+V + +E QH FN P+Q A P ++D + D P +
Sbjct 242 VVCRSQEQQHQFNTPYQLASPPPGYDFDA------------------------VSDGPES 277
Query 64 IQSTSVPVQDGDLIILGTDGLFDNV---FDFEVLALTGLAVSPHEAQTLLGDASLATPPE 120
+ P+Q GD+I+L TDG++DNV F EVL +P Q
Sbjct 278 ADTIQFPMQLGDVILLATDGVYDNVPESFLVEVLTEMSGISNPVRLQM------------ 325
Query 121 DVARALALAAYWRSLDRDAQTPFSKEARR 149
A +AL A S +PFS+ AR+
Sbjct 326 -AANTVALMARTLSFSPKHDSPFSQNARK 353
> CE16565
Length=330
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/146 (29%), Positives = 61/146 (41%), Gaps = 39/146 (26%)
Query 4 LVKKVKEMQHGFNVPFQFAHLPAPEEWDGLLQKGLTRLVGIAKQEYGQGNGGKIGDEPSA 63
+V K +E H FN PFQ LP PE + G IGD+
Sbjct 201 IVSKSREQVHYFNAPFQLT-LP-PEGYQGF-----------------------IGDKADM 235
Query 64 IQSTSVPVQDGDLIILGTDGLFDNVFDFEVL-ALTGLAVSPHEAQTLLGDASLATPPEDV 122
+ V+ GD+I+L TDG++DN+ + +VL L L Q +V
Sbjct 236 ADKDEMAVKKGDIILLATDGVWDNLSEQQVLDQLKALDAGKSNVQ-------------EV 282
Query 123 ARALALAAYWRSLDRDAQTPFSKEAR 148
ALAL A + D +PF+ +AR
Sbjct 283 CNALALTARRLAFDSKHNSPFAMKAR 308
> SPAC1556.03
Length=288
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 62/130 (47%), Gaps = 22/130 (16%)
Query 23 HLPAPEEWDGLLQKGLTRLVGIAKQEY--GQGNGGKIGDEPSAIQSTSVPVQDGDLIILG 80
H +P + +LQ + + I + Y + G K+G Q+T ++D DL+IL
Sbjct 161 HYASPAQ---VLQFNMPYQLAIYPRNYRSAENIGPKMG------QATVHDLKDNDLVILA 211
Query 81 TDGLFDNVFDFEVLALTGLA--VSPHEAQTLLGDASLATPPEDVARALALAAYWRSLDRD 138
TDG+FDN+ + +L + G+ S Q L +D+A + A SLD
Sbjct 212 TDGIFDNIEEKSILDIAGVVDFSSLSNVQKCL---------DDLAMRICRQAVLNSLDTK 262
Query 139 AQTPFSKEAR 148
++PF+K A+
Sbjct 263 WESPFAKTAK 272
> 7301827
Length=314
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 41/149 (27%), Positives = 68/149 (45%), Gaps = 36/149 (24%)
Query 4 LVKKVKEMQHGFNVPFQFAHLPAPEEWDGLLQKGLTRLVGIAKQEYGQGNGGKIGDEPSA 63
++ + E H FN P+Q +P PE+ K+ Y D+P
Sbjct 178 VLHRSVEQTHDFNTPYQLT-VP-PED---------------RKESY-------YCDKPEM 213
Query 64 IQSTSVPVQDGDLIILGTDGLFDNVFDFEVLALTGLAVSPHEAQTLLGDASLATPPEDVA 123
ST + GDL++L TDGLFDN+ + +L++ E L+G + + + A
Sbjct 214 AVSTRHSLLPGDLVLLATDGLFDNMPESMLLSILNGLKERGEHDLLVGASRVV----EKA 269
Query 124 RALALAAYWRSLDRDAQTPFSKEARRCHN 152
R L++ A + Q+PF+ +AR+ HN
Sbjct 270 RELSMNASF-------QSPFAIKARQ-HN 290
> At4g33500_1
Length=702
Score = 39.7 bits (91), Expect = 0.004, Method: Composition-based stats.
Identities = 21/80 (26%), Positives = 42/80 (52%), Gaps = 12/80 (15%)
Query 69 VPVQDGDLIILGTDGLFDNVFDFEVLALTGLAVSPHEAQTLLGDASLATPPEDVARALAL 128
V +++GD++I TDGLFDN+++ E++++ + G + P+ +A +A
Sbjct 613 VNLEEGDVVIAATDGLFDNLYEKEIVSI------------VCGSLKQSLEPQKIAELVAA 660
Query 129 AAYWRSLDRDAQTPFSKEAR 148
A + +TPF+ A+
Sbjct 661 KAQEVGRSKTERTPFADAAK 680
> YHR076w
Length=374
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 49/199 (24%), Positives = 71/199 (35%), Gaps = 63/199 (31%)
Query 4 LVKKVKEMQHGFNVPFQFAHLPAPEEWDGLLQKGLTRLVGIAKQEYGQGNGGK-IGDEPS 62
LV + K GFN P+Q + +P EE +L++ R G K I + P
Sbjct 235 LVFQTKFQTVGFNAPYQLSIIP--EE---MLKEAERR-------------GSKYILNTPR 276
Query 63 AIQSTSVPVQDGDLIILGTDGLFDNVFDFEVLALTGLAVSPHEAQTLLGDASLATPPEDV 122
S ++ D+IIL TDG+ DN ++ + + L D + T E
Sbjct 277 DADEYSFQLKKKDIIILATDGVTDN-------------IATDDIELFLKDNAARTNDELQ 323
Query 123 ARALALAAYWRSLDRDAQTP--FSKEARRCHNKQSLPVQTPDGSSEQGAPQASPVHAMFL 180
+ SL +D P F++E + K
Sbjct 324 LLSQKFVDNVVSLSKDPNYPSVFAQEISKLTGKN-------------------------- 357
Query 181 AGLLSGGKEDDITVAAAWV 199
SGGKEDDITV V
Sbjct 358 ---YSGGKEDDITVVVVRV 373
> Hs21361256
Length=299
Score = 33.5 bits (75), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 36/71 (50%), Gaps = 7/71 (9%)
Query 75 DLIILGTDGLFDNVFDFEVLALTGLAVSPHEAQTLLGDASLATPPEDVARALALAAYWRS 134
D+++LGTDGL+D D EV A +S +E D S T +A+AL L A
Sbjct 215 DVLVLGTDGLWDVTTDCEVAATVDRVLSAYEPN----DHSRYTA---LAQALVLGARGTP 267
Query 135 LDRDAQTPFSK 145
DR + P +K
Sbjct 268 RDRGWRLPNNK 278
> At1g76430
Length=532
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 43/113 (38%), Gaps = 19/113 (16%)
Query 105 EAQTLLGDASLATPPEDVARAL---------ALAAYWR----------SLDRDAQTPFSK 145
E + G +LA P D+A L AL YWR +L + +K
Sbjct 192 EKTNVAGLETLAPPESDIAWRLILMIGALPAALTFYWRMLMPETARYTALVENNVVQAAK 251
Query 146 EARRCHNKQSLPVQTPDGSSEQGAPQASPVHAMFLAGLLSGGKEDDITVAAAW 198
+ +R + + T D SSE P +S + +F LS D +A W
Sbjct 252 DMQRVMSVSMISQITEDSSSELEQPPSSSSYKLFSRRFLSLHGRDLFAASANW 304
> SPCC1739.03
Length=1015
Score = 30.0 bits (66), Expect = 3.2, Method: Composition-based stats.
Identities = 28/106 (26%), Positives = 42/106 (39%), Gaps = 23/106 (21%)
Query 66 STSVPVQDGDLIILGTDGLFDNVFDFEVL--------ALTGLAVSPHEAQTLLGDASLAT 117
+TS + G L++L D DF+ L+GL PHE D S++
Sbjct 234 ATSRRLISGSLVLLSND-------DFQTFRIGTVCARPLSGLNKHPHEIDVKFEDISISL 286
Query 118 PP-EDVARALALAAYW-------RSLDRDAQTPFSKEARRCHNKQS 155
P E+ A + YW RSL R + + F + H K +
Sbjct 287 DPREEYVMIEATSGYWEAYKHVLRSLQRLSASTFPMKDYLVHCKSN 332
> Hs22045740
Length=628
Score = 29.6 bits (65), Expect = 3.9, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 1/45 (2%)
Query 2 LSLVKKVKEMQHGFNVPFQFAHLPAPEEWDGLLQKGLTRLVGIAK 46
LS K E+ G P+Q+ HL PE+WDG + LT+ I K
Sbjct 99 LSDPKLAMEVFFGPCTPYQY-HLHGPEKWDGARRANLTQRERIIK 142
> At5g58900
Length=288
Score = 29.3 bits (64), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 152 NKQSLPVQTPDGSSEQGAPQASPVHAMFLAGLLSGGKEDDITVAAAWVCRK 202
NK+S ++P+ ++G P H +FL GL GK D ++ +V +
Sbjct 124 NKRSQAGRSPELERKKGVPWTEEEHKLFLMGLKKYGKGDWRNISRNFVITR 174
Lambda K H
0.317 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3682951018
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40