bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0030_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
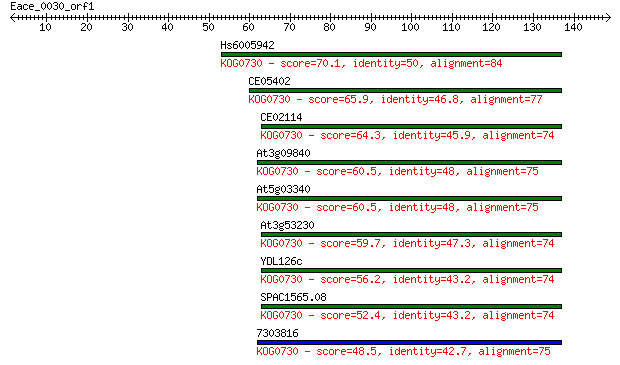
Hs6005942 70.1 1e-12
CE05402 65.9 3e-11
CE02114 64.3 7e-11
At3g09840 60.5 1e-09
At5g03340 60.5 1e-09
At3g53230 59.7 2e-09
YDL126c 56.2 2e-08
SPAC1565.08 52.4 3e-07
7303816 48.5 4e-06
> Hs6005942
Length=806
Score = 70.1 bits (170), Expect = 1e-12, Method: Composition-based stats.
Identities = 42/91 (46%), Positives = 56/91 (61%), Gaps = 11/91 (12%)
Query 53 ISVDPWGEYR---RVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFTFRG- 108
IS+ P + + R+H+LP DT+ + LFE YL+P+ E +R IRKGDIF RG
Sbjct 100 ISIQPCPDVKYGKRIHVLPIDDTVEGI-TGNLFEVYLKPYFLEA-YRPIRKGDIFLVRGG 157
Query 109 ---VEFKVIATEPSTVPVARVGPETVVHFEG 136
VEFKV+ T+PS P V P+TV+H EG
Sbjct 158 MRAVEFKVVETDPS--PYCIVAPDTVIHCEG 186
> CE05402
Length=810
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 36/82 (43%), Positives = 53/82 (64%), Gaps = 9/82 (10%)
Query 60 EY-RRVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFT----FRGVEFKVI 114
EY +RVH+LP DT+ + LF+ +L+P+ + +R + KGDIFT R VEFKV+
Sbjct 114 EYGKRVHVLPIDDTI-EGLTGNLFDVFLRPYFTDA-YRPVHKGDIFTVQAAMRTVEFKVV 171
Query 115 ATEPSTVPVARVGPETVVHFEG 136
T+P+ P V P+TV+H+EG
Sbjct 172 ETDPA--PACIVAPDTVIHYEG 191
> CE02114
Length=809
Score = 64.3 bits (155), Expect = 7e-11, Method: Composition-based stats.
Identities = 34/78 (43%), Positives = 50/78 (64%), Gaps = 8/78 (10%)
Query 63 RVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFT----FRGVEFKVIATEP 118
R+H+LP DT+ + LF+ +L+P+ E +R + KGDIFT R VEFKV+ TEP
Sbjct 119 RIHVLPIDDTI-EGLTGNLFDVFLKPYFLEA-YRPLHKGDIFTVQAAMRTVEFKVVETEP 176
Query 119 STVPVARVGPETVVHFEG 136
+ P V P+T++H+EG
Sbjct 177 A--PACIVSPDTMIHYEG 192
> At3g09840
Length=809
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 36/79 (45%), Positives = 49/79 (62%), Gaps = 8/79 (10%)
Query 62 RRVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFTFRG----VEFKVIATE 117
+RVHILP DT+ + LF+ YL+P+ E +R +RKGD+F RG VEFKVI T+
Sbjct 116 KRVHILPVDDTVEGV-TGNLFDAYLKPYFLEA-YRPVRKGDLFLVRGGMRSVEFKVIETD 173
Query 118 PSTVPVARVGPETVVHFEG 136
P+ V V P+T + EG
Sbjct 174 PAEYCV--VAPDTEIFCEG 190
> At5g03340
Length=843
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 36/79 (45%), Positives = 49/79 (62%), Gaps = 8/79 (10%)
Query 62 RRVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFTFRG----VEFKVIATE 117
+RVHILP DT+ + LF+ YL+P+ E +R +RKGD+F RG VEFKVI T+
Sbjct 149 KRVHILPVDDTVEGV-TGNLFDAYLKPYFLEA-YRPVRKGDLFLVRGGMRSVEFKVIETD 206
Query 118 PSTVPVARVGPETVVHFEG 136
P+ V V P+T + EG
Sbjct 207 PAEYCV--VAPDTEIFCEG 223
> At3g53230
Length=815
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 35/78 (44%), Positives = 48/78 (61%), Gaps = 8/78 (10%)
Query 63 RVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFTFRG----VEFKVIATEP 118
RVHILP DT+ S +F+ YL+P+ E +R +RKGD+F RG +EFKVI T+P
Sbjct 118 RVHILPLDDTIEGV-SGNIFDAYLKPYFLEA-YRPVRKGDLFLVRGGMRSIEFKVIETDP 175
Query 119 STVPVARVGPETVVHFEG 136
+ V V P+T + EG
Sbjct 176 AEYCV--VAPDTEIFCEG 191
> YDL126c
Length=835
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 32/78 (41%), Positives = 47/78 (60%), Gaps = 8/78 (10%)
Query 63 RVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFTFRG----VEFKVIATEP 118
R+ +LP ADT+ + LF+ +L+P+ E +R +RKGD F RG VEFKV+ EP
Sbjct 123 RISVLPIADTIEGI-TGNLFDVFLKPYFVEA-YRPVRKGDHFVVRGGMRQVEFKVVDVEP 180
Query 119 STVPVARVGPETVVHFEG 136
V V +T++H+EG
Sbjct 181 EEYAV--VAQDTIIHWEG 196
> SPAC1565.08
Length=418
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/78 (41%), Positives = 47/78 (60%), Gaps = 8/78 (10%)
Query 63 RVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFTFRG----VEFKVIATEP 118
R+ +LP ADT+ + LF+ YL+P+ E +R IRKGD+F RG VEFKV+ P
Sbjct 133 RISVLPLADTV-EGLTGSLFDVYLKPYFVEA-YRPIRKGDLFVVRGSMRQVEFKVVDVAP 190
Query 119 STVPVARVGPETVVHFEG 136
+ V +T++H+EG
Sbjct 191 DEFGI--VSQDTIIHWEG 206
> 7303816
Length=801
Score = 48.5 bits (114), Expect = 4e-06, Method: Composition-based stats.
Identities = 32/79 (40%), Positives = 42/79 (53%), Gaps = 8/79 (10%)
Query 62 RRVHILPFADTLPTTYSFRLFEDYLQPFLKEQPWRLIRKGDIFTFRG----VEFKVIATE 117
+RV ILP D + LFE YL+P+ E +R I GD F R +EFKV+ T+
Sbjct 109 KRVRILPI-DESTEGVTGNLFEIYLKPYFLEA-YRPIHMGDNFIVRAAMRPIEFKVVLTD 166
Query 118 PSTVPVARVGPETVVHFEG 136
P P V PETV+ +G
Sbjct 167 PE--PYCIVAPETVIFCDG 183
Lambda K H
0.324 0.141 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40