bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0031_orf1
Length=192
Score E
Sequences producing significant alignments: (Bits) Value
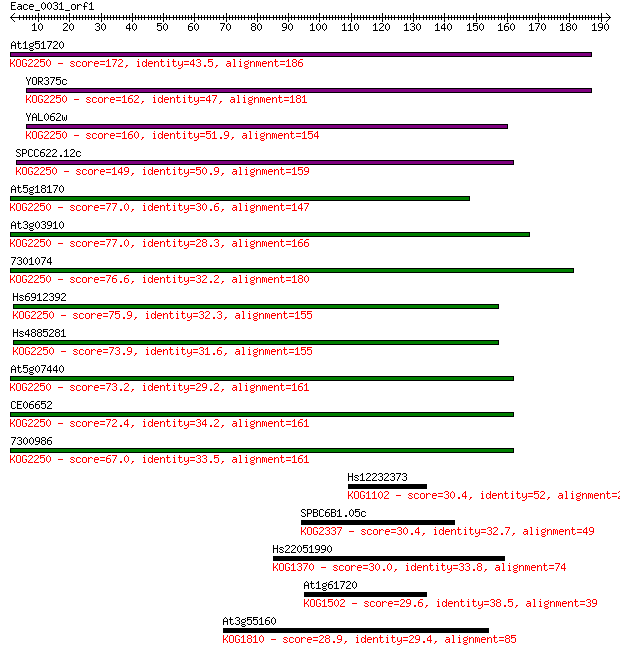
At1g51720 172 4e-43
YOR375c 162 5e-40
YAL062w 160 2e-39
SPCC622.12c 149 4e-36
At5g18170 77.0 2e-14
At3g03910 77.0 2e-14
7301074 76.6 2e-14
Hs6912392 75.9 5e-14
Hs4885281 73.9 2e-13
At5g07440 73.2 3e-13
CE06652 72.4 5e-13
7300986 67.0 2e-11
Hs12232373 30.4 2.2
SPBC6B1.05c 30.4 2.6
Hs22051990 30.0 3.3
At1g61720 29.6 4.0
At3g55160 28.9 6.0
> At1g51720
Length=624
Score = 172 bits (436), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 81/186 (43%), Positives = 122/186 (65%), Gaps = 2/186 (1%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GGSDFDPKGKS EI RFCQS M E+ +++GP +D+P+ ++GVGTRE+G+LFGQY+RL
Sbjct 301 GGSDFDPKGKSDNEIMRFCQSFMNEMYRYMGPDKDLPSEEVGVGTREMGYLFGQYRRLAG 360
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
F+G+ TG I W S +R EA+GYG YFA +L + +KG +C++SGCG +A++
Sbjct 361 QFQGSFTGPRIYWAASSLRTEASGYGVVYFARLILAD-MNKEIKGLRCVVSGCGKIAMHV 419
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVVFER 180
EKL+ GA +T+SDS G + GF ++ +++ K+ R++R + + F+
Sbjct 420 VEKLIACGAHPVTVSDSKGYLVDDDGFDYMKLAFLRDIKSQQRSLRDYSKTYARAKYFDE 479
Query 181 RGTPWS 186
PW+
Sbjct 480 L-KPWN 484
> YOR375c
Length=454
Score = 162 bits (409), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 85/189 (44%), Positives = 117/189 (61%), Gaps = 10/189 (5%)
Query 6 DPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCSNFEGA 65
D KG+S EIRR C + M EL++HIG DVPAGDIGVG REIG+LFG Y+ +++EG
Sbjct 116 DLKGRSNNEIRRICYAFMRELSRHIGQDTDVPAGDIGVGGREIGYLFGAYRSYKNSWEGV 175
Query 66 LTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLY--DTLKGKKCILSGCGNVAVYCAEK 123
LTGK + WGGS IRPEATGYG Y+ + ++D ++ +GK+ +SG GNVA Y A K
Sbjct 176 LTGKGLNWGGSLIRPEATGYGLVYYTQAMIDYATNGKESFEGKRVTISGSGNVAQYAALK 235
Query 124 LLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRVEAFANGESVVFERR-- 181
++E+G V++LSDS G I G + E++ I AK ++ +E N S E +
Sbjct 236 VIELGGTVVSLSDSKGCIISETGITSEQVADISSAKVNFKS--LEQIVNEYSTFSENKVQ 293
Query 182 ----GTPWS 186
PW+
Sbjct 294 YIAGARPWT 302
> YAL062w
Length=457
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 80/156 (51%), Positives = 106/156 (67%), Gaps = 2/156 (1%)
Query 6 DPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCSNFEGA 65
D KGKS EIRR C + M EL++HIG DVPAGDIGVG REIG+LFG Y+ +++EG
Sbjct 117 DLKGKSDNEIRRICYAFMRELSRHIGKDTDVPAGDIGVGGREIGYLFGAYRSYKNSWEGV 176
Query 66 LTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLY--DTLKGKKCILSGCGNVAVYCAEK 123
LTGK + WGGS IRPEATG+G Y+ + ++D ++ +GK+ +SG GNVA Y A K
Sbjct 177 LTGKGLNWGGSLIRPEATGFGLVYYTQAMIDYATNGKESFEGKRVTISGSGNVAQYAALK 236
Query 124 LLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAK 159
++E+G V++LSDS G I G + E+I I AK
Sbjct 237 VIELGGIVVSLSDSKGCIISETGITSEQIHDIASAK 272
> SPCC622.12c
Length=451
Score = 149 bits (375), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 81/166 (48%), Positives = 109/166 (65%), Gaps = 7/166 (4%)
Query 3 SDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCSNF 62
SDFDPKGKS EIRRF Q+ M +L ++IGP DVPAGDIGV + +FG+YKRL + +
Sbjct 116 SDFDPKGKSDNEIRRFSQAFMRQLFRYIGPQTDVPAGDIGVTGFVVMHMFGEYKRLRNEY 175
Query 63 EGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLY--DTLKGKKCILSGCGNVAVYC 120
G +TGK + GGS IRPEATGYG Y+ +++++ +TLKGK+ +SG GNVA Y
Sbjct 176 SGVVTGKHMLTGGSNIRPEATGYGVVYYVKHMIEHRTKGAETLKGKRVAISGSGNVAQYA 235
Query 121 AEKLLEMGAEVLTLSDSSGTIYC--SAGFSKEEIRTI---KEAKAT 161
A K ++ GA V ++SDS G + + G EEI I KE +A+
Sbjct 236 ALKCIQEGAIVKSISDSKGVLIAKTAEGLVPEEIHEIMALKEKRAS 281
> At5g18170
Length=411
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/147 (30%), Positives = 76/147 (51%), Gaps = 1/147 (0%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GG DP S++E+ R + ++ IG DVPA D+G G + + ++ +Y +
Sbjct 103 GGIGCDPSKLSISELERLTRVFTQKIHDLIGIHTDVPAPDMGTGPQTMAWILDEYSKFHG 162
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
+TGK I GGS R ATG G + E +L++ T+ G++ ++ G GNV +
Sbjct 163 YSPAVVTGKPIDLGGSLGRDAATGRGVMFGTEALLNE-HGKTISGQRFVIQGFGNVGSWA 221
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGF 147
A+ + E G +++ +SD +G I G
Sbjct 222 AKLISEKGGKIVAVSDITGAIKNKDGI 248
> At3g03910
Length=411
Score = 77.0 bits (188), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 47/166 (28%), Positives = 86/166 (51%), Gaps = 4/166 (2%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GG DP S++E+ R + ++ IG DVPA D+G G + + ++ +Y +
Sbjct 103 GGIGCDPSELSLSELERLTRVFTQKIHDLIGIHTDVPAPDMGTGPQTMAWILDEYSKFHG 162
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
+ +TGK I GGS R ATG G + E +L++ T+ G++ + G GNV +
Sbjct 163 HSPAVVTGKPIDLGGSLGRDAATGRGVLFATEALLNE-HGKTISGQRFAIQGFGNVGSWA 221
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVR 166
A+ + + G +++ +SD +G I + G +I ++ E +R ++
Sbjct 222 AKLISDKGGKIVAVSDVTGAIKNNNGI---DILSLLEHAEENRGIK 264
> 7301074
Length=590
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 58/193 (30%), Positives = 87/193 (45%), Gaps = 13/193 (6%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKR- 57
G +PK S E+ + + ELAK IGP DVPA D+G G RE+ ++ Y +
Sbjct 177 AGLKINPKEYSEHELEKITRRFTLELAKKGFIGPGVDVPAPDMGTGEREMSWIADTYAKT 236
Query 58 ---LCSNFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDT-------LKGKK 107
L N +TGK I GG R ATG G + EN +++ Y + GK
Sbjct 237 IGHLDINAHACVTGKPINQGGIHGRVSATGRGVFHGLENFINEANYMSQIGTTPGWGGKT 296
Query 108 CILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKATDRAVRV 167
I+ G GNV ++ L GA + + + GT+Y G + + K T +
Sbjct 297 FIVQGFGNVGLHTTRYLTRAGATCIGVIEHDGTLYNPEGIDPKLLEDYKNEHGTIVGYQN 356
Query 168 EAFANGESVVFER 180
GE+++FE+
Sbjct 357 AKPYEGENLMFEK 369
> Hs6912392
Length=558
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 50/168 (29%), Positives = 80/168 (47%), Gaps = 13/168 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +PK + E+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 185 GVKINPKNYTENELEKITRRFTMELAKKGFIGPGVDVPAPDMNTGEREMSWIADTYASTI 244
Query 60 SNFE----GALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
+++ +TGK I GG R ATG G + EN ++Q Y ++ G K
Sbjct 245 GHYDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFINQASYMSILGMTPGFRDKTF 304
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIK 156
++ G GNV ++ L GA+ + + +S G+I+ G +E+ K
Sbjct 305 VVQGFGNVGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKELEDFK 352
> Hs4885281
Length=558
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 49/168 (29%), Positives = 80/168 (47%), Gaps = 13/168 (7%)
Query 2 GSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRLC 59
G +PK + E+ + + ELAK IGP DVPA D+ G RE+ ++ Y
Sbjct 185 GVKINPKNYTDNELEKITRRFTMELAKKGFIGPGIDVPAPDMSTGEREMSWIADTYASTI 244
Query 60 SNFE----GALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKG-------KKC 108
+++ +TGK I GG R ATG G + EN +++ Y ++ G K
Sbjct 245 GHYDINAHACVTGKPISQGGIHGRISATGRGVFHGIENFINEASYMSILGMTPGFGDKTF 304
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIK 156
++ G GNV ++ L GA+ + + +S G+I+ G +E+ K
Sbjct 305 VVQGFGNVGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKELEDFK 352
> At5g07440
Length=411
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 47/161 (29%), Positives = 79/161 (49%), Gaps = 3/161 (1%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKHIGPSRDVPAGDIGVGTREIGFLFGQYKRLCS 60
GG P+ S++E+ R + ++ IG DVPA D+G + + ++ +Y +
Sbjct 103 GGIGCSPRDLSLSELERLTRVFTQKIHDLIGIHTDVPAPDMGTNAQTMAWILDEYSKFHG 162
Query 61 NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYC 120
+ +TGK I GGS R ATG G + E +L + +++G ++ G GNV +
Sbjct 163 HSPAVVTGKPIDLGGSLGREAATGRGVVFATEALLAE-YGKSIQGLTFVIQGFGNVGTWA 221
Query 121 AEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
A+ + E G +V+ +SD +G I G + IK AT
Sbjct 222 AKLIHEKGGKVVAVSDITGAIRNPEGIDINAL--IKHKDAT 260
> CE06652
Length=536
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 55/174 (31%), Positives = 80/174 (45%), Gaps = 13/174 (7%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRL 58
GG DPK + EI + + E AK +GP DVPA D+G G RE+G++ Y +
Sbjct 162 GGVKIDPKQYTDYEIEKITRRIAIEFAKKGFLGPGVDVPAPDMGTGEREMGWIADTYAQT 221
Query 59 CSNFE----GALTGKDIGWGGSEIRPEATGYGCAYFAENVLDQLLY------DT-LKGKK 107
+ + +TGK I GG R ATG G E + Y DT L GK
Sbjct 222 IGHLDRDASACITGKPIVSGGIHGRVSATGRGVWKGLEVFTNDADYMKMVGLDTGLAGKT 281
Query 108 CILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
I+ G GNV ++ L G++V+ + + +Y G +E+ K+A T
Sbjct 282 AIIQGFGNVGLHTHRYLHRAGSKVIGIQEYDCAVYNPDGIHPKELEDWKDANGT 335
> 7300986
Length=520
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 54/174 (31%), Positives = 80/174 (45%), Gaps = 13/174 (7%)
Query 1 GGSDFDPKGKSVAEIRRFCQSSMTELAKH--IGPSRDVPAGDIGVGTREIGFLFGQYKRL 58
GG DPK +V E++ + EL K IGP DVPA D+ G RE+ ++ QY++
Sbjct 148 GGICIDPKKYTVDELQTITRRYTMELLKRNMIGPGIDVPAPDVNTGPREMSWIVDQYQKT 207
Query 59 CS----NFEGALTGKDIGWGGSEIRPEATGYGCAYFAENVL-DQLLYDTL------KGKK 107
N +TGK + GG R ATG G + L D+ D L K K
Sbjct 208 FGYKDINSSAIVTGKPVHNGGINGRHSATGRGVWKAGDLFLKDKEWMDLLKWKTGWKDKT 267
Query 108 CILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIYCSAGFSKEEIRTIKEAKAT 161
I+ G GNV + A+ + E GA+V+ + + ++ G ++ E K T
Sbjct 268 VIVQGFGNVGSFAAKYVHEAGAKVIGIKEFDVSLVNKDGIDINDLFEYTEEKKT 321
> Hs12232373
Length=997
Score = 30.4 bits (67), Expect = 2.2, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 109 ILSGCGNVAVYCAEKLLEMGAEVLT 133
+LSG G+V+ CAEK+LE E+L+
Sbjct 450 LLSGSGDVSKECAEKILETWGELLS 474
> SPBC6B1.05c
Length=649
Score = 30.4 bits (67), Expect = 2.6, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 94 VLDQLLYDTLKGKKCILSGCGNVAVYCAEKLLEMGAEVLTLSDSSGTIY 142
++ QL D ++ KC+L G G + A LL G +T D S Y
Sbjct 324 LVPQLDLDRIQNSKCLLLGAGTLGCGVARNLLSWGVRHVTFVDYSTVSY 372
> Hs22051990
Length=648
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 39/78 (50%), Gaps = 9/78 (11%)
Query 85 YGCAYFAENVLDQLLYDT---LKGKKCILSGCGNVAVYCAEKLLEMGAEV-LTLSDSSGT 140
YGC E+++D + + T + GK +++G GNV CA+ L GA V +T D
Sbjct 504 YGCW---ESLIDGIKWATVVMIAGKVAMVAGYGNVGKGCAQALWGFGAHVIITKIDPINA 560
Query 141 IYCSAGFSKEEIRTIKEA 158
+ A E+ T+ EA
Sbjct 561 L--QAAMEGYEVTTMDEA 576
> At1g61720
Length=340
Score = 29.6 bits (65), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 95 LDQLLYDTLKGKKCILSGCGNVAVYCAEKLLEMGAEVLT 133
+DQ L T K C++ G GN+A + LL+ G +V T
Sbjct 1 MDQTLTHTGSKKACVIGGTGNLASILIKHLLQSGYKVNT 39
> At3g55160
Length=2149
Score = 28.9 bits (63), Expect = 6.0, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 37/85 (43%), Gaps = 12/85 (14%)
Query 69 KDIGWGGSEIRPEATGYGCAYFAENVLDQLLYDTLKGKKCILSGCGNVAVYCAEKLLEMG 128
+D WG E G A + E+ L LY GK + S N+ Y + LLE+
Sbjct 523 RDESWGS-----EGVDQGYARYREHCLPPFLYGLASGKSKLRS---NLNTYAVQVLLELD 574
Query 129 AEVLTLSDSSGTIYCSAGFSKEEIR 153
+ + L + Y S G S+EE +
Sbjct 575 VDSIFLLLA----YISIGPSEEETK 595
Lambda K H
0.318 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3238438140
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40