bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0045_orf2
Length=154
Score E
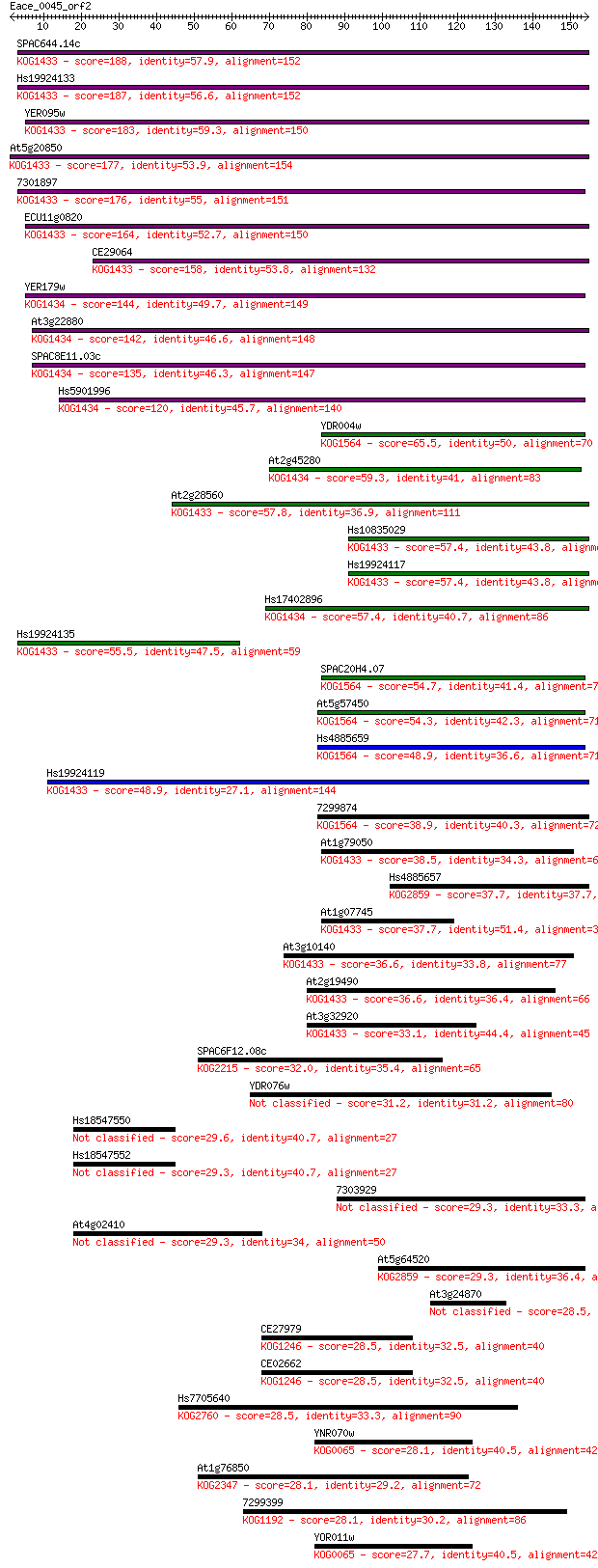
Sequences producing significant alignments: (Bits) Value
SPAC644.14c 188 4e-48
Hs19924133 187 7e-48
YER095w 183 1e-46
At5g20850 177 9e-45
7301897 176 1e-44
ECU11g0820 164 7e-41
CE29064 158 3e-39
YER179w 144 6e-35
At3g22880 142 2e-34
SPAC8E11.03c 135 4e-32
Hs5901996 120 8e-28
YDR004w 65.5 4e-11
At2g45280 59.3 3e-09
At2g28560 57.8 7e-09
Hs10835029 57.4 1e-08
Hs19924117 57.4 1e-08
Hs17402896 57.4 1e-08
Hs19924135 55.5 5e-08
SPAC20H4.07 54.7 7e-08
At5g57450 54.3 9e-08
Hs4885659 48.9 4e-06
Hs19924119 48.9 4e-06
7299874 38.9 0.004
At1g79050 38.5 0.006
Hs4885657 37.7 0.009
At1g07745 37.7 0.010
At3g10140 36.6 0.018
At2g19490 36.6 0.019
At3g32920 33.1 0.20
SPAC6F12.08c 32.0 0.43
YDR076w 31.2 0.87
Hs18547550 29.6 2.6
Hs18547552 29.3 2.8
7303929 29.3 2.9
At4g02410 29.3 3.0
At5g64520 29.3 3.1
At3g24870 28.5 4.7
CE27979 28.5 5.3
CE02662 28.5 5.3
Hs7705640 28.5 6.1
YNR070w 28.1 6.1
At1g76850 28.1 7.0
7299399 28.1 7.6
YOR011w 27.7 9.7
> SPAC644.14c
Length=365
Score = 188 bits (477), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 88/152 (57%), Positives = 115/152 (75%), Gaps = 0/152 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
GP+ L+ L G+T D+ + E G +TVE +A+ P + LL IKGISE KA KL + +
Sbjct 43 GPMPLQMLEGNGITASDIKKIHEAGYYTVESIAYTPKRQLLLIKGISEAKADKLLGEASK 102
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
L +GF +A EY R+ LI TTGS QLD+LL+GG+ETG++TELFGEFRTGK+Q+CHTL
Sbjct 103 LVPMGFTTATEYHIRRSELITITTGSKQLDTLLQGGVETGSITELFGEFRTGKSQICHTL 162
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
AV+CQLP++ GGEGKCL+IDTEGTFRP R++
Sbjct 163 AVTCQLPIDMGGGEGKCLYIDTEGTFRPVRLL 194
> Hs19924133
Length=339
Score = 187 bits (475), Expect = 7e-48, Method: Compositional matrix adjust.
Identities = 86/152 (56%), Positives = 116/152 (76%), Gaps = 0/152 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+ + +
Sbjct 21 GPQPISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKILAEAAK 80
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
L +GF +A E+ + R+ +I+ TTGS +LD LL+GGIETG++TE+FGEFRTGKTQ+CHTL
Sbjct 81 LVPMGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQICHTL 140
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
AV+CQLP+++ GGEGK ++IDTEGTFRPER++
Sbjct 141 AVTCQLPIDRGGGEGKAMYIDTEGTFRPERLL 172
> YER095w
Length=400
Score = 183 bits (465), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 89/150 (59%), Positives = 112/150 (74%), Gaps = 0/150 (0%)
Query 5 LKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELC 64
+ +E L G+T D+ L+E GLHT E VA+AP K LL IKGISE KA KL + L
Sbjct 81 VPIEKLQVNGITMADVKKLRESGLHTAEAVAYAPRKDLLEIKGISEAKADKLLNEAARLV 140
Query 65 SLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAV 124
+GF +A ++ R+ LI TTGS LD+LL GG+ETG++TELFGEFRTGK+QLCHTLAV
Sbjct 141 PMGFVTAADFHMRRSELICLTTGSKNLDTLLGGGVETGSITELFGEFRTGKSQLCHTLAV 200
Query 125 SCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
+CQ+P++ GGEGKCL+IDTEGTFRP R+V
Sbjct 201 TCQIPLDIGGGEGKCLYIDTEGTFRPVRLV 230
> At5g20850
Length=339
Score = 177 bits (448), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 83/154 (53%), Positives = 113/154 (73%), Gaps = 0/154 (0%)
Query 1 QTGPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVS 60
Q GP +E L A G+ D+ L++ GL TVE VA+ P K LL IKGIS+ K K+ + +
Sbjct 19 QHGPFPVEQLQAAGIASVDVKKLRDAGLCTVEGVAYTPRKDLLQIKGISDAKVDKIVEAA 78
Query 61 KELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCH 120
+L LGF SA + R +I+ T+GS +LD +L+GGIETG++TEL+GEFR+GKTQLCH
Sbjct 79 SKLVPLGFTSASQLHAQRQEIIQITSGSRELDKVLEGGIETGSITELYGEFRSGKTQLCH 138
Query 121 TLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
TL V+CQLP++Q GGEGK ++ID EGTFRP+R++
Sbjct 139 TLCVTCQLPMDQGGGEGKAMYIDAEGTFRPQRLL 172
> 7301897
Length=336
Score = 176 bits (447), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 83/151 (54%), Positives = 112/151 (74%), Gaps = 0/151 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKE 62
GPL + L+ +T KD+ LL++ LHTVE VA A K L+AI G+ K ++ + +
Sbjct 18 GPLSVTKLIGGSITAKDIKLLQQASLHTVESVANATKKQLMAIPGLGGGKVEQIITEANK 77
Query 63 LCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTL 122
L LGF SA+ + + RA++++ +TGS +LD LL GGIETG++TE+FGEFR GKTQLCHTL
Sbjct 78 LVPLGFLSARTFYQMRADVVQLSTGSKELDKLLGGGIETGSITEIFGEFRCGKTQLCHTL 137
Query 123 AVSCQLPVEQSGGEGKCLWIDTEGTFRPERI 153
AV+CQLP+ Q GGEGKC++IDTE TFRPER+
Sbjct 138 AVTCQLPISQKGGEGKCMYIDTENTFRPERL 168
> ECU11g0820
Length=334
Score = 164 bits (415), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 79/150 (52%), Positives = 106/150 (70%), Gaps = 0/150 (0%)
Query 5 LKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELC 64
+ + L G+ D+ L E G TVE +AFAP + LL+IKG S+ K KL + + +L
Sbjct 17 ISISELKNGGILAVDIAKLIEAGFTTVESLAFAPKRQLLSIKGFSDIKVDKLIKEAAKLV 76
Query 65 SLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAV 124
+GF +A Y + R+ L+ TTGS ++D LL GG E+G++TE+FGEFRTGKTQLCHT+AV
Sbjct 77 PMGFTTASAYHQRRSELVYLTTGSSEVDKLLSGGFESGSITEIFGEFRTGKTQLCHTVAV 136
Query 125 SCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
+CQLP EQ GG GK ++IDTEGTFR ER+V
Sbjct 137 TCQLPPEQGGGGGKAMYIDTEGTFRSERLV 166
> CE29064
Length=395
Score = 158 bits (400), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 71/132 (53%), Positives = 96/132 (72%), Gaps = 0/132 (0%)
Query 23 LKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLGFCSAQEYLEARANLI 82
LKE G +T E +AF + L +KGIS+QKA K+ + + + +GF + E R+ L+
Sbjct 95 LKEAGYYTYESLAFTTRRELRNVKGISDQKAEKIMKEAMKFVQMGFTTGAEVHVKRSQLV 154
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWI 142
+ TGS LD LL GGIETG++TE++GE+RTGKTQLCH+LAV CQLP++ GGEGKC++I
Sbjct 155 QIRTGSASLDRLLGGGIETGSITEVYGEYRTGKTQLCHSLAVLCQLPIDMGGGEGKCMYI 214
Query 143 DTEGTFRPERIV 154
DT TFRPERI+
Sbjct 215 DTNATFRPERII 226
> YER179w
Length=334
Score = 144 bits (363), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 74/149 (49%), Positives = 97/149 (65%), Gaps = 0/149 (0%)
Query 5 LKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELC 64
L ++ L G+ DL LK GG++TV V + L IKG+SE K K+K+ + ++
Sbjct 17 LSVDELQNYGINASDLQKLKSGGIYTVNTVLSTTRRHLCKIKGLSEVKVEKIKEAAGKII 76
Query 65 SLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAV 124
+GF A L+ R + +TGS QLDS+L GGI T ++TE+FGEFR GKTQ+ HTL V
Sbjct 77 QVGFIPATVQLDIRQRVYSLSTGSKQLDSILGGGIMTMSITEVFGEFRCGKTQMSHTLCV 136
Query 125 SCQLPVEQSGGEGKCLWIDTEGTFRPERI 153
+ QLP E GGEGK +IDTEGTFRPERI
Sbjct 137 TTQLPREMGGGEGKVAYIDTEGTFRPERI 165
> At3g22880
Length=339
Score = 142 bits (358), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 69/148 (46%), Positives = 97/148 (65%), Gaps = 0/148 (0%)
Query 7 LEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSL 66
++ L+A+G+ D+ L+E G+HT + K L IKG+SE K K+ + ++++ +
Sbjct 31 IDKLIAQGINAGDVKKLQEAGIHTCNGLMMHTKKNLTGIKGLSEAKVDKICEAAEKIVNF 90
Query 67 GFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSC 126
G+ + + L R +++K TTG LD LL GGIET +TE FGEFR+GKTQL HTL V+
Sbjct 91 GYMTGSDALIKRKSVVKITTGCQALDDLLGGGIETSAITEAFGEFRSGKTQLAHTLCVTT 150
Query 127 QLPVEQSGGEGKCLWIDTEGTFRPERIV 154
QLP GG GK +IDTEGTFRP+RIV
Sbjct 151 QLPTNMKGGNGKVAYIDTEGTFRPDRIV 178
> SPAC8E11.03c
Length=332
Score = 135 bits (339), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 68/147 (46%), Positives = 96/147 (65%), Gaps = 0/147 (0%)
Query 7 LEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSL 66
+E L A G+ D+ LK+ G+ TV+ V + + LL IKG SE K KLK+ + ++C
Sbjct 18 IEDLTAHGIGMTDIIKLKQAGVCTVQGVHMSTKRFLLKIKGFSEAKVDKLKEAASKMCPA 77
Query 67 GFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSC 126
F +A E + R + +TGS L+ +L GGI++ ++TE+FGEFR GKTQ+ HTL V+
Sbjct 78 NFSTAMEISQNRKKVWSISTGSEALNGILGGGIQSMSITEVFGEFRCGKTQMSHTLCVTA 137
Query 127 QLPVEQSGGEGKCLWIDTEGTFRPERI 153
QLP + G EGK +IDTEGTFRP+RI
Sbjct 138 QLPRDMGGAEGKVAFIDTEGTFRPDRI 164
> Hs5901996
Length=340
Score = 120 bits (302), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 64/140 (45%), Positives = 86/140 (61%), Gaps = 0/140 (0%)
Query 14 GLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLGFCSAQE 73
G+ D LK G+ T++ + + L +KG+SE K K+K+ + +L GF +A E
Sbjct 31 GINVADNKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVDKIKEAANKLIEPGFLTAFE 90
Query 74 YLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQS 133
Y E R + TTGS + D LL GGIE+ +TE FGEFRTGKTQL HTL V+ QLP
Sbjct 91 YSEKRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTAQLPGAGG 150
Query 134 GGEGKCLWIDTEGTFRPERI 153
GK ++IDTE TFRP+R+
Sbjct 151 YPGGKIIFIDTENTFRPDRL 170
> YDR004w
Length=460
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 35/70 (50%), Positives = 44/70 (62%), Gaps = 0/70 (0%)
Query 84 FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWID 143
FTT V +D LL GGI T +TE+FGE TGK+QL LA+S QL G GKC++I
Sbjct 100 FTTADVAMDELLGGGIFTHGITEIFGESSTGKSQLLMQLALSVQLSEPAGGLGGKCVYIT 159
Query 144 TEGTFRPERI 153
TEG +R+
Sbjct 160 TEGDLPTQRL 169
> At2g45280
Length=332
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 50/83 (60%), Gaps = 0/83 (0%)
Query 70 SAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLP 129
+A + L +L + TT LD++L GGI ++TE+ G GKTQ+ L+V+ Q+P
Sbjct 61 NAWDMLHEEESLPRITTSCSDLDNILGGGISCRDVTEIGGVPGIGKTQIGIQLSVNVQIP 120
Query 130 VEQSGGEGKCLWIDTEGTFRPER 152
E G GK ++IDTEG+F ER
Sbjct 121 RECGGLGGKAIYIDTEGSFMVER 143
> At2g28560
Length=353
Score = 57.8 bits (138), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 41/111 (36%), Positives = 52/111 (46%), Gaps = 7/111 (6%)
Query 44 AIKGISEQKAAKLKQVSKELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGN 103
AI ISE + + VS SL F E +L G LD L GGI G
Sbjct 52 AISFISEATSPPCQSVS----SLFFFKKVENEHLSGHLPTHLKG---LDDTLCGGIPFGV 104
Query 104 LTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
LTEL G GK+Q C LA+S PV G +G+ ++ID E F R++
Sbjct 105 LTELVGPPGIGKSQFCMKLALSASFPVAYGGLDGRVIYIDVESKFSSRRVI 155
> Hs10835029
Length=350
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 91 LDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRP 150
LD L GG+ G+LTE+ G GKTQ C +++ LP G EG ++IDTE F
Sbjct 90 LDEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTESAFSA 149
Query 151 ERIV 154
ER+V
Sbjct 150 ERLV 153
> Hs19924117
Length=350
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/64 (43%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 91 LDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRP 150
LD L GG+ G+LTE+ G GKTQ C +++ LP G EG ++IDTE F
Sbjct 90 LDEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTESAFSA 149
Query 151 ERIV 154
ER+V
Sbjct 150 ERLV 153
> Hs17402896
Length=376
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 35/86 (40%), Positives = 46/86 (53%), Gaps = 0/86 (0%)
Query 69 CSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQL 128
C+A E LE T LD +L GG+ TE+ G GKTQLC LAV Q+
Sbjct 85 CTALELLEQEHTQGFIITFCSALDDILGGGVPLMKTTEICGAPGVGKTQLCMQLAVDVQI 144
Query 129 PVEQSGGEGKCLWIDTEGTFRPERIV 154
P G G+ ++IDTEG+F +R+V
Sbjct 145 PECFGGVAGEAVFIDTEGSFMVDRVV 170
> Hs19924135
Length=242
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/59 (47%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 3 GPLKLEHLLAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSK 61
GP + L G+ D+ L+E G HTVE VA+AP K L+ IKGISE KA K+ V++
Sbjct 21 GPQPISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKILAVAE 79
> SPAC20H4.07
Length=354
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 39/70 (55%), Gaps = 0/70 (0%)
Query 84 FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWID 143
TTG V+LD L GGI G LTE+ GE +GK+Q C L + QLP+ G ++I
Sbjct 75 LTTGDVKLDETLHGGIPVGQLTEICGESGSGKSQFCMQLCLMVQLPLSLGGMNKAAVFIS 134
Query 144 TEGTFRPERI 153
TE +R+
Sbjct 135 TESGLETKRL 144
> At5g57450
Length=304
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 40/71 (56%), Gaps = 0/71 (0%)
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWI 142
K TTG LD L+GGI +LTE+ E GKTQLC L++ QLP+ G G L++
Sbjct 20 KLTTGCEILDGCLRGGISCDSLTEIVAESGCGKTQLCLQLSLCTQLPISHGGLNGSSLYL 79
Query 143 DTEGTFRPERI 153
+E F R+
Sbjct 80 HSEFPFPFRRL 90
> Hs4885659
Length=346
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 39/71 (54%), Gaps = 0/71 (0%)
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWI 142
+ + G LD+LL+GG+ +TEL G GKTQL L ++ Q P + G E ++I
Sbjct 81 RLSLGCPVLDALLRGGLPLDGITELAGRSSAGKTQLALQLCLAVQFPRQHGGLEAGAVYI 140
Query 143 DTEGTFRPERI 153
TE F +R+
Sbjct 141 CTEDAFPHKRL 151
> Hs19924119
Length=328
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 39/145 (26%), Positives = 72/145 (49%), Gaps = 6/145 (4%)
Query 11 LAKGLTKKDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVS-KELCSLGFC 69
L GLT++ + LL+ + TV + A L+ + G+S + L++V + +
Sbjct 8 LCPGLTEEMIQLLRSHRIKTVVDLVSADLEEVAQKCGLSYKALVALRRVLLAQFSAFPVN 67
Query 70 SAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLP 129
A + E + + +TG LD LL G+ TG +TE+ G +GKTQ+C +A +
Sbjct 68 GADLHEELKTSTAILSTGIGSLDKLLDAGLYTGEVTEIVGGPGSGKTQVCLCMAANVAHG 127
Query 130 VEQSGGEGKCLWIDTEGTFRPERIV 154
++Q+ L++D+ G R++
Sbjct 128 LQQN-----VLYVDSNGGLTASRLL 147
> 7299874
Length=341
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 39/73 (53%), Gaps = 2/73 (2%)
Query 83 KFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGK-CLW 141
+ + G LD GG+ T +TEL G GKTQL L++ QLP E GG GK +
Sbjct 87 RVSFGCSALDRCTGGGVVTRGITELCGAAGVGKTQLLLQLSLCVQLPRE-LGGLGKGVAY 145
Query 142 IDTEGTFRPERIV 154
I TE +F R++
Sbjct 146 ICTESSFPARRLL 158
> At1g79050
Length=432
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 36/67 (53%), Gaps = 6/67 (8%)
Query 84 FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWID 143
F++G + LD L GG+ G + E++G +GKT TLA+ V++ G G + +D
Sbjct 117 FSSGILTLDLALGGGLPKGRVVEIYGPESSGKT----TLALHAIAEVQKLG--GNAMLVD 170
Query 144 TEGTFRP 150
E F P
Sbjct 171 AEHAFDP 177
> Hs4885657
Length=280
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 102 GNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGTFRPERIV 154
G++ E G TGKT++ + L C LP + G E + L+IDT+ F R+V
Sbjct 41 GDILEFHGPEGTGKTEMLYHLTARCILPKSEGGLEVEVLFIDTDYHFDMLRLV 93
> At1g07745
Length=200
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 84 FTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQL 118
+TG + DSLL+GG G LTEL G +GKTQ+
Sbjct 67 LSTGDKETDSLLQGGFREGQLTELVGPSSSGKTQM 101
> At3g10140
Length=389
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 43/78 (55%), Gaps = 9/78 (11%)
Query 74 YLEARANLIKFTTGSVQLD-SLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQ 132
Y + R ++I +TGS+ LD +L GG+ G + E++G+ +GKT TLA+ ++
Sbjct 89 YRKRRVSVI--STGSLNLDLALGVGGLPKGRMVEVYGKEASGKT----TLALHIIKEAQK 142
Query 133 SGGEGKCLWIDTEGTFRP 150
G G C ++D E P
Sbjct 143 LG--GYCAYLDAENAMDP 158
> At2g19490
Length=376
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 37/67 (55%), Gaps = 7/67 (10%)
Query 80 NLIKFTTGSVQLDSLLK-GGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGK 138
N+ F+TGS LD L GG+ G + E++G +GKT TLA+ ++ G G
Sbjct 35 NVPVFSTGSFALDVALGVGGLPKGRVVEIYGPEASGKT----TLALHVIAEAQKQG--GT 88
Query 139 CLWIDTE 145
C+++D E
Sbjct 89 CVFVDAE 95
> At3g32920
Length=229
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 28/47 (59%), Gaps = 2/47 (4%)
Query 80 NLIKFTTGSVQLDSLLK-GGIETGNLTELFGEFRTGKTQLC-HTLAV 124
N+ F+TGS LD L GG+ G L E++G +GKT L H L++
Sbjct 11 NVPVFSTGSFALDVALGVGGLPKGRLVEIYGPEASGKTALALHMLSM 57
> SPAC6F12.08c
Length=578
Score = 32.0 bits (71), Expect = 0.43, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 37/65 (56%), Gaps = 6/65 (9%)
Query 51 QKAAKLKQVSKELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGE 110
Q AK + + KE+ S+ F ++L+ + +L +FT L +L++ +E GNL ELF E
Sbjct 353 QAYAKQRNMVKEI-SMQF----DHLDIQISLQEFTDAIRNL-ALIQNKLERGNLNELFAE 406
Query 111 FRTGK 115
F K
Sbjct 407 FFADK 411
> YDR076w
Length=406
Score = 31.2 bits (69), Expect = 0.87, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 40/81 (49%), Gaps = 4/81 (4%)
Query 65 SLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAV 124
SLG +Q +E+ L ++G LD +L G + ++ E+FG GKT L
Sbjct 2 SLGIPLSQLIVESPKPL---SSGITGLDEILNLGFQARSIYEIFGPPGIGKTNFGIQLVC 58
Query 125 SCQLPVEQSG-GEGKCLWIDT 144
+ ++QS + K LWI+T
Sbjct 59 NSLEGIQQSEINDDKILWIET 79
> Hs18547550
Length=138
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 18 KDLDLLKEGGLHTVECVAFAPLKTLLA 44
D++L +E LHT+ C+A +P LL+
Sbjct 111 SDINLFREPFLHTLSCIAASPTSCLLS 137
> Hs18547552
Length=138
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 18 KDLDLLKEGGLHTVECVAFAPLKTLLA 44
D++L +E LHT+ C+A +P LL+
Sbjct 111 SDINLFREPFLHTLSCIAASPTSCLLS 137
> 7303929
Length=184
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 5/66 (7%)
Query 88 SVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGGEGKCLWIDTEGT 147
S+ + K + G + EL G+ GKTQL +TLA++ Q+ L+IDT+
Sbjct 7 SITIQESKKQPFKPGRVWELCGQPGVGKTQLLYTLALNFVWKHSQA-----VLFIDTKRE 61
Query 148 FRPERI 153
F +RI
Sbjct 62 FSCKRI 67
> At4g02410
Length=674
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 0/50 (0%)
Query 18 KDLDLLKEGGLHTVECVAFAPLKTLLAIKGISEQKAAKLKQVSKELCSLG 67
KD DLL GG V K +A+K +S + LK+ E+ S+G
Sbjct 356 KDKDLLGSGGFGRVYRGVMPTTKKEIAVKRVSNESRQGLKEFVAEIVSIG 405
> At5g64520
Length=403
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 5/60 (8%)
Query 99 IETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQS----GGEGK-CLWIDTEGTFRPERI 153
+ GN+ E+ G + KTQ+ A+SC LP + GG GK L++D + F R+
Sbjct 40 LRAGNVVEITGASTSAKTQILIQAAISCILPKTWNGIHYGGLGKLVLFLDLDCRFDVLRL 99
> At3g24870
Length=1841
Score = 28.5 bits (62), Expect = 4.7, Method: Composition-based stats.
Identities = 10/20 (50%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 113 TGKTQLCHTLAVSCQLPVEQ 132
T TQ+CH +A++CQL E+
Sbjct 594 TAATQICHRVALTCQLRFEE 613
> CE27979
Length=1013
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 68 FCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTEL 107
+C++QE LEA L++F +V D L+ + L +L
Sbjct 541 YCTSQEVLEAAGQLLEFHVNAVYKDERLQPPVADKTLLDL 580
> CE02662
Length=972
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 68 FCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTEL 107
+C++QE LEA L++F +V D L+ + L +L
Sbjct 500 YCTSQEVLEAAGQLLEFHVNAVYKDERLQPPVADKTLLDL 539
> Hs7705640
Length=386
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 42/99 (42%), Gaps = 17/99 (17%)
Query 46 KGISE--QKAAKLKQVSKELCSLGFCSAQEYLEARANLIKFTTGSVQLDS-------LLK 96
K ISE + +KL +KE+ L ++ AN IK G + D LL+
Sbjct 172 KNISEAFEDLSKLMIKAKEMVELS--------KSIANKIKDKQGDITEDETIRFKSYLLR 223
Query 97 GGIETGNLTELFGEFRTGKTQLCHTLAVSCQLPVEQSGG 135
GI E +G QL LA Q+P+E+ GG
Sbjct 224 MGIANPVTRETYGSGTQYHMQLAKQLAGILQVPLEERGG 262
> YNR070w
Length=1333
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 82 IKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLA 123
I ++G +L + G G LT L GE GKT L +TLA
Sbjct 738 IPHSSGQRKLLDSVSGYCVPGTLTALIGESGAGKTTLLNTLA 779
> At1g76850
Length=1085
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Query 51 QKAAKLKQVSKELCSLGFCSAQEYLEARANLIKFTTGSVQLDSLLKGGIETGNLTELFGE 110
+ ++ L+ + LG + Y A+ANLI+ + LDS GI+ G+ ++ G
Sbjct 821 EDSSDLQDLIMSFSGLGEKVLEHYTFAKANLIRTAATNYLLDS----GIQWGSAPQVKG- 875
Query 111 FRTGKTQLCHTL 122
R +L HTL
Sbjct 876 IRDAAVELLHTL 887
> 7299399
Length=519
Score = 28.1 bits (61), Expect = 7.6, Method: Composition-based stats.
Identities = 26/88 (29%), Positives = 37/88 (42%), Gaps = 7/88 (7%)
Query 63 LCSLGFCSAQEYLEA--RANLIKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCH 120
LCSLG+ S YL A F G +L KG ++ G+ + F K +
Sbjct 11 LCSLGYSSGYNYLMVLNSAGRSHFNVGH----ALAKGLVKAGHTITVVSVFPQKKPIPGY 66
Query 121 TLAVSCQLPVEQSGGEGKCLWIDTEGTF 148
T VS +E GG+ LW + T+
Sbjct 67 T-DVSVPNVIEVMGGDIGALWASIQKTY 93
> YOR011w
Length=1394
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 24/42 (57%), Gaps = 1/42 (2%)
Query 82 IKFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLA 123
I +T G+ +L + G I +G LT L GE GKT L + L+
Sbjct 756 INYTVGTKKLINNASGFISSG-LTALMGESGAGKTTLLNVLS 796
Lambda K H
0.317 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40