bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0054_orf1
Length=285
Score E
Sequences producing significant alignments: (Bits) Value
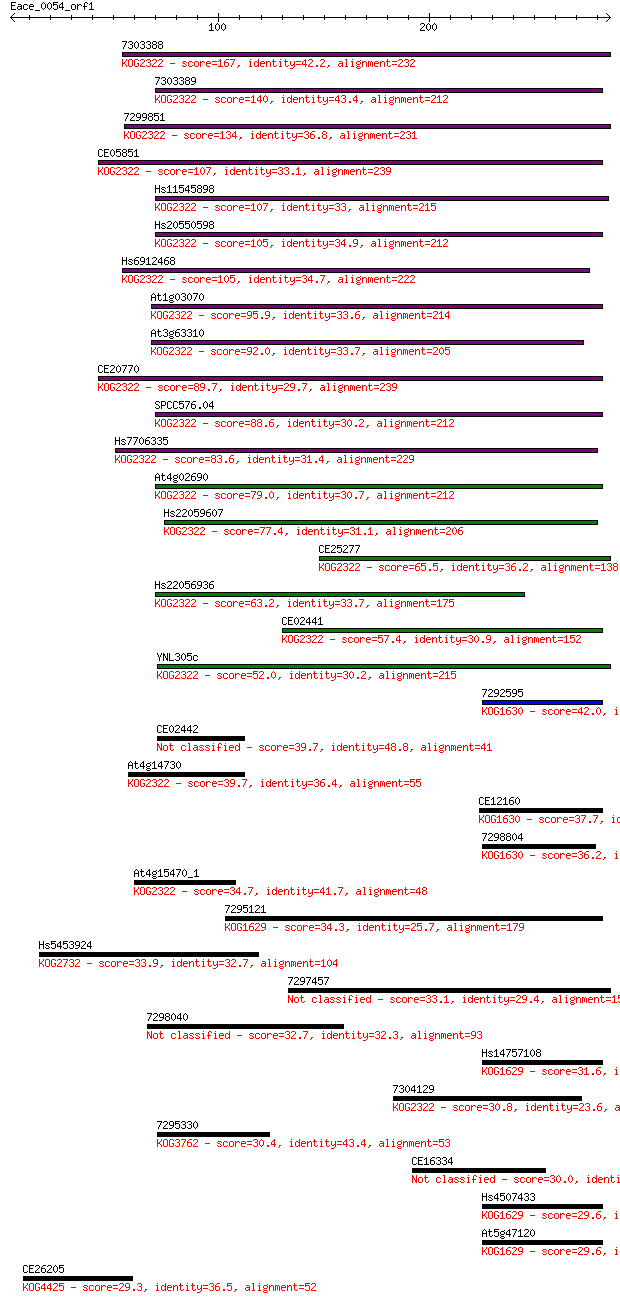
7303388 167 3e-41
7303389 140 3e-33
7299851 134 2e-31
CE05851 107 2e-23
Hs11545898 107 3e-23
Hs20550598 105 8e-23
Hs6912468 105 1e-22
At1g03070 95.9 8e-20
At3g63310 92.0 1e-18
CE20770 89.7 6e-18
SPCC576.04 88.6 1e-17
Hs7706335 83.6 4e-16
At4g02690 79.0 1e-14
Hs22059607 77.4 3e-14
CE25277 65.5 1e-10
Hs22056936 63.2 6e-10
CE02441 57.4 3e-08
YNL305c 52.0 2e-06
7292595 42.0 0.002
CE02442 39.7 0.007
At4g14730 39.7 0.007
CE12160 37.7 0.029
7298804 36.2 0.079
At4g15470_1 34.7 0.25
7295121 34.3 0.28
Hs5453924 33.9 0.35
7297457 33.1 0.73
7298040 32.7 0.90
Hs14757108 31.6 2.2
7304129 30.8 3.1
7295330 30.4 4.0
CE16334 30.0 6.6
Hs4507433 29.6 7.1
At5g47120 29.6 7.3
CE26205 29.3 9.1
> 7303388
Length=324
Score = 167 bits (422), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 98/242 (40%), Positives = 135/242 (55%), Gaps = 11/242 (4%)
Query 54 PPPNGNGLQISEDISVQ----------IRHAFVRKVLGILAIQILFTFGIAAVFGFVPTL 103
PPP+ G +D Q IR F+RKV IL Q++ TFG A+F +
Sbjct 82 PPPSAGGYGAYDDPESQPKNFSFDDQSIRRGFIRKVYLILMGQLIVTFGAVALFVYHEGT 141
Query 104 RNFLLQNYWLAIVAAVCALVLQLVLVCVPDLARKVPTNFILMSLITACYSVLISCAAAAS 163
+ F N WL VA LV L + C + R+ PTNFI + L TA S L+ +A
Sbjct 142 KTFARNNMWLFWVALGVMLVTMLSMACCESVRRQTPTNFIFLGLFTAAQSFLMGVSATKY 201
Query 164 SYSSFLIAIGATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLMIFGILCIIFSSKVVH 223
+ L+A+G T V + L +FA QTKYDFT G L ++ +IFGI+ I K++
Sbjct 202 APKEVLMAVGITAAVCLALTIFALQTKYDFTMMGGILIACMVVFLIFGIVAIFVKGKIIT 261
Query 224 LVYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAIAD 283
LVY+ + +LF + L+YDTQ ++GG+H +Y S ++YIFAAL LY+DII IFM IL I
Sbjct 262 LVYASIGALLFSVYLIYDTQLMMGGEH-KYSISPEEYIFAALNLYLDIINIFMYILTIIG 320
Query 284 NS 285
S
Sbjct 321 AS 322
> 7303389
Length=239
Score = 140 bits (353), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 92/212 (43%), Positives = 123/212 (58%), Gaps = 1/212 (0%)
Query 70 QIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLVLV 129
IR F+RKV IL Q+L TFG +VF F + ++ +N L +A +V + +
Sbjct 23 SIRKGFIRKVYLILMCQLLITFGFVSVFTFSKASQEWVQKNPALFWIALAVLIVTMICMA 82
Query 130 CVPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLFACQT 189
C + RK P NFI + L T S L+ A L+A+G T V +GL LFA QT
Sbjct 83 CCESVRRKTPLNFIFLFLFTVAESFLLGMVAGQFEADEVLMAVGITAAVALGLTLFALQT 142
Query 190 KYDFTGWGTYLFVAVLCLMIFGILCIIFSSKVVHLVYSGLATVLFCMILVYDTQQVVGGK 249
KYDFT G L ++ +IFGI+ I KV+ LVY+ L +LF + LVYDTQ ++GG
Sbjct 143 KYDFTMCGGVLVACLVVFIIFGIIAIFIPGKVIGLVYASLGALLFSVYLVYDTQLMLGGN 202
Query 250 HRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
H +Y S ++YIFAAL LY+DII IFM IL I
Sbjct 203 H-KYSISPEEYIFAALNLYLDIINIFMYILTI 233
> 7299851
Length=264
Score = 134 bits (337), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 85/231 (36%), Positives = 127/231 (54%), Gaps = 6/231 (2%)
Query 55 PPNGNGLQISEDISVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLA 114
P G G S IR F+RKV IL Q++ + + +R + ++ W+
Sbjct 39 PDKGLGF-----CSASIRRGFIRKVYLILLAQLITSLVVIVSLTADKRVRLMVAESTWIF 93
Query 115 IVAAVCALVLQLVLVCVPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGA 174
+VA + + + L C DL R+ P NFI +S T S L+ AA + +A+
Sbjct 94 VVAILIVVFSLVALGCNEDLRRQTPANFIFLSAFTIAESFLLGVAACRYAPMEIFMAVLI 153
Query 175 TFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLMIFGILCIIFSSKVVHLVYSGLATVLF 234
T V +GL LFA QT+YDFT G L ++ L+ FGI+ I +V +Y+ L+ +LF
Sbjct 154 TASVCLGLTLFALQTRYDFTVMGGLLVSCLIILLFFGIVTIFVGGHMVTTIYASLSALLF 213
Query 235 CMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAIADNS 285
+ LVYDTQ ++GGKH RY S ++YIFAAL +YMD++ IF++IL + S
Sbjct 214 SVYLVYDTQLMMGGKH-RYSISPEEYIFAALNIYMDVMNIFLDILQLIGGS 263
> CE05851
Length=244
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 79/248 (31%), Positives = 131/248 (52%), Gaps = 18/248 (7%)
Query 43 IESAQHKQVSSPPPNGN-GLQISEDISVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVP 101
+E + P+G L S S +R AFVRKV ++ I F I A F +P
Sbjct 1 MEQGYGATTAQDDPDGKYNLHFS---SQTVRAAFVRKVFMLVTIM----FAITAAFCVIP 53
Query 102 TL----RNFLLQNYWLAIVAAVCALVLQLVLVCVPDLARKVPTNFILMSLITACYSVLIS 157
+ ++++ N+W+ +A + LV+ + L C +L R+ P N IL+++ T +V+
Sbjct 54 MVSEPFQDWVKNNFWVYFIAIIVFLVVAIALSCCGNLRRQFPVNIILLTIFTLSAAVMTM 113
Query 158 CAAAASSYSSFLIAIGATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLMIFGILCIIF 217
A + S LI + T V +++F+ +TK D T F+ + L FGI +IF
Sbjct 114 FVTACYNVQSVLICLCITTVCSGSVIIFSMKTKSDLTSKMGIAFMLSMVLFSFGIFALIF 173
Query 218 S----SKVVHLVYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIII 273
+ + ++ VYSGLA +L L D Q ++GG R+Y+ S +DYIFAA+ +++DI+
Sbjct 174 TLAFNWQFLYSVYSGLAALLMMFYLAIDVQLLMGG--RKYELSPEDYIFAAMEIFLDILN 231
Query 274 IFMNILAI 281
IF+ +L I
Sbjct 232 IFLMLLNI 239
> Hs11545898
Length=311
Score = 107 bits (266), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 71/219 (32%), Positives = 120/219 (54%), Gaps = 6/219 (2%)
Query 70 QIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLVLV 129
++RH F+RKV I+++Q+L T I A+F FV + F+ +N + V+ +V L+L
Sbjct 93 KVRHTFIRKVYSIISVQLLITVAIIAIFTFVEPVSAFVRRNVAVYYVSYAVFVVTYLILA 152
Query 130 CVPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLFACQT 189
C R+ P N IL++L T + ++ + +IA+ T VV + + +F QT
Sbjct 153 CCQGPRRRFPWNIILLTLFTFAMGFMTGTISSMYQTKAVIIAMIITAVVSISVTIFCFQT 212
Query 190 KYDFTGWGTYLFVAVLCLMIFGILC--IIFSSKV--VHLVYSGLATVLFCMILVYDTQQV 245
K DFT V + L++ GI+ +++ V +H++Y+ L + F + L YDTQ V
Sbjct 213 KVDFTSCTGLFCVLGIVLLVTGIVTSIVLYFQYVYWLHMLYAALGAICFTLFLAYDTQLV 272
Query 246 VGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAIADN 284
+G +R++ S +DYI AL +Y DII IF +L + +
Sbjct 273 LG--NRKHTISPEDYITGALQIYTDIIYIFTFVLQLMGD 309
> Hs20550598
Length=316
Score = 105 bits (263), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 74/219 (33%), Positives = 117/219 (53%), Gaps = 12/219 (5%)
Query 70 QIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQN---YWLAIVAAVCALVLQL 126
++R FVRKV IL IQ+L T + A+F F +++++ N YW + L
Sbjct 97 KVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPVKDYVQANPGWYW---ASYAVFFATYL 153
Query 127 VLVCVPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLFA 186
L C R P N IL+++ T + L ++ + +S L+ +G T +V + + +F+
Sbjct 154 TLACCSGPRRHFPWNLILLTVFTLSMAYLTGMLSSYYNTTSVLLCLGITALVCLSVTVFS 213
Query 187 CQTKYDFTGWGTYLFVAVLCLMIFG-ILCIIFSSKVV---HLVYSGLATVLFCMILVYDT 242
QTK+DFT LFV ++ L G IL I+ + V H VY+ L +F + L DT
Sbjct 214 FQTKFDFTSCQGVLFVLLMTLFFSGLILAILLPFQYVPWLHAVYAALGAGVFTLFLALDT 273
Query 243 QQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
Q ++G +RR+ S ++YIF AL +Y+DII IF L +
Sbjct 274 QLLMG--NRRHSLSPEEYIFGALNIYLDIIYIFTFFLQL 310
> Hs6912468
Length=316
Score = 105 bits (261), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 77/227 (33%), Positives = 118/227 (51%), Gaps = 8/227 (3%)
Query 54 PPPNGNGLQISEDISVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLL-QNYW 112
PP + +S ++ ++R FVRKV IL IQ+L T + A+F F + Q W
Sbjct 81 PPETMSSSPLSAGMTKKVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPCQGLCSGQPGW 140
Query 113 LAIVAAVCALVLQLVLVCVPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAI 172
AV L L C R P N IL+++ T + L ++ + +S L+ +
Sbjct 141 YWASYAVF-FATYLTLACCSGPRRHFPWNLILLTVFTLSMAYLTGMLSSYYNTTSVLLCL 199
Query 173 GATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLMIFG-ILCIIFSSKVV---HLVYSG 228
G T +V + + +F+ QTK+DFT LFV ++ L G IL I+ + V H VY+
Sbjct 200 GITALVCLSVTVFSFQTKFDFTSCQGVLFVLLMTLFFSGLILAILLPFQYVPWLHAVYAA 259
Query 229 LATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIF 275
L +F + L DTQ ++G +RR+ S ++YIF AL +Y+DII IF
Sbjct 260 LGAGVFTLFLALDTQLLMG--NRRHSLSPEEYIFGALNIYLDIIYIF 304
> At1g03070
Length=247
Score = 95.9 bits (237), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 72/226 (31%), Positives = 119/226 (52%), Gaps = 26/226 (11%)
Query 68 SVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLV 127
S ++R F+RKV I+A Q+L T +A+ FV + F L L +V
Sbjct 31 SPELRWGFIRKVYSIIAFQLLATIAVASTVVFVRPIAVFF--------ATTSAGLALWIV 82
Query 128 LVCVPDLA--------RKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVV 179
L+ P + +K P N++L+ + T + + A +S L A T VVV
Sbjct 83 LIITPLIVMCPLYYYHQKHPVNYLLLGIFTVALAFAVGLTCAFTSGKVILEAAILTTVVV 142
Query 180 VGLMLF---ACQTKYDFTGWGTYLFVAVLCLMIFGILCIIFS-SKVVHLVYSGLATVLFC 235
+ L ++ A + YDF G +LF A++ LM+F ++ I F ++ ++Y LA ++FC
Sbjct 143 LSLTVYTFWAAKKGYDFNFLGPFLFGALIVLMVFALIQIFFPLGRISVMIYGCLAAIIFC 202
Query 236 MILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
+VYDT ++ +RY Y D+YI+AA++LY+DII +F+ +L I
Sbjct 203 GYIVYDTDNLI----KRYSY--DEYIWAAVSLYLDIINLFLALLTI 242
> At3g63310
Length=239
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 69/211 (32%), Positives = 118/211 (55%), Gaps = 14/211 (6%)
Query 68 SVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLV 127
S ++R +F+RKV I++IQ+L T +AA V ++ F A A L+L +
Sbjct 22 SPELRWSFIRKVYSIISIQLLVTIAVAATVVKVHSISVFFTTT--TAGFALYILLILTPL 79
Query 128 LVCVP--DLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLF 185
+V P +K P N++L+ + T + + A +S L ++ T VVV+ L L+
Sbjct 80 IVMCPLYYYHQKHPVNYLLLGIFTVALAFAVGLTCAFTSGKVILESVILTAVVVISLTLY 139
Query 186 ---ACQTKYDFTGWGTYLFVAVLCLMIFGILCIIFS-SKVVHLVYSGLATVLFCMILVYD 241
A + +DF G +LF AV+ LM+F + I+F K+ ++Y LA+++FC +VYD
Sbjct 140 TFWAAKRGHDFNFLGPFLFGAVIVLMVFSFIQILFPLGKISVMIYGCLASIIFCGYIVYD 199
Query 242 TQQVVGGKHRRYQYSIDDYIFAALTLYMDII 272
T ++ +R+ Y D+YI+AA++LY+D+I
Sbjct 200 TDNLI----KRHSY--DEYIWAAVSLYLDVI 224
> CE20770
Length=295
Score = 89.7 bits (221), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 71/248 (28%), Positives = 123/248 (49%), Gaps = 20/248 (8%)
Query 43 IESAQHKQVSSPPPNGN-GLQISEDISVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVP 101
+E+ QH P+G Q S+ +R AFVRKV ++ I + + +
Sbjct 55 MENGQHG--GGDNPDGKYSFQFSDK---TVRAAFVRKVFSLVFIMLCIVAAVTVIPWVHD 109
Query 102 TLRNFLLQNYWLAIVAAVCALVLQLVLVCVPDLARKVPTNFILMSLITACYSVLISCAAA 161
+ +N L + + V V L LVC + RK P N I+ + T SV+ +A
Sbjct 110 DTMRMVRRNSALYLGSYVIFFVTYLSLVCCEGVRRKFPVNLIVTGIFTLATSVMTMVISA 169
Query 162 ASSYSSFLIA----IGATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLMIFGILCII- 216
+ L+A IG TF +V+ A QTK+D T Y+ + +C M FG++ +I
Sbjct 170 HHDANVVLLALAICIGCTFSIVI----VASQTKFDLTAHMGYILIISMCFMFFGLVVVIC 225
Query 217 ---FSSKVVHLVYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIII 273
F K + +VY+ ++ + L D Q ++GGK +Y+ S ++YIFA++ +++DI+
Sbjct 226 SMFFKIKFLMMVYALGGALIMMLYLFLDVQMLMGGK--KYEISPEEYIFASVQIFIDIVQ 283
Query 274 IFMNILAI 281
+F +L++
Sbjct 284 MFWFLLSL 291
> SPCC576.04
Length=266
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 64/213 (30%), Positives = 109/213 (51%), Gaps = 10/213 (4%)
Query 70 QIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLVLV 129
IR AF+RKV IL Q+ T +F P ++ + W I+ +LV+ L+
Sbjct 53 SIRMAFLRKVYAILTAQLFVTSLFGGIFYLHPAFSFWVQMHPWFLILNFFISLVVLFGLI 112
Query 130 CVPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLFACQT 189
P P N+I + L TA + + A S L A+ T V V L F Q+
Sbjct 113 MKP---YSYPRNYIFLFLFTALEGLTLGTAITFFSARIILEAVFITLGVFVALTAFTFQS 169
Query 190 KYDFTGWGTYLFVAVLCLMIFG-ILCIIFSSKVVHLVYSGLATVLFCMILVYDTQQVVGG 248
K+DF+ G +L+V++ L++ I + S+ + + ++G T++FC +++DT ++
Sbjct 170 KWDFSRLGGFLYVSLWSLILTPLIFFFVPSTPFIDMAFAGFGTLVFCGYILFDTYNIL-- 227
Query 249 KHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
HR YS +++I ++L LY+D I +F+ IL I
Sbjct 228 -HR---YSPEEFIMSSLMLYLDFINLFIRILQI 256
> Hs7706335
Length=258
Score = 83.6 bits (205), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 72/241 (29%), Positives = 126/241 (52%), Gaps = 24/241 (9%)
Query 51 VSSPPPNGNGLQISEDI---------SVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVP 101
++ P P I +D +V IR AF+RKV IL++Q+L T + VF +
Sbjct 21 MADPDPRYPRSSIEDDFNYGSSVASATVHIRMAFLRKVYSILSLQVLLTTVTSTVFLYFE 80
Query 102 TLRNFLLQNYWLAIVAAVCALVLQLVLVCVPDLAR-KVPTNFILMSLITACYSVLISCAA 160
++R F+ ++ L ++ A+ +L L L L R K P N L+ T + ++ A
Sbjct 81 SVRTFVHESPALILLFALGSLGLIFALT----LNRHKYPLNLYLLFGFTLLEA--LTVAV 134
Query 161 AASSYSSFLI--AIGATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLMIFGILCIIFS 218
+ Y ++I A T V GL ++ Q+K DF+ +G LF + L + G L F
Sbjct 135 VVTFYDVYIILQAFILTTTVFFGLTVYTLQSKKDFSKFGAGLFALLWILCLSGFLKFFFY 194
Query 219 SKVVHLVYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNI 278
S+++ LV + +LFC ++YDT ++ ++ S ++Y+ AA++LY+DII +F+++
Sbjct 195 SEIMELVLAAAGALLFCGFIIYDTHSLM------HKLSPEEYVLAAISLYLDIINLFLHL 248
Query 279 L 279
L
Sbjct 249 L 249
> At4g02690
Length=248
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 65/217 (29%), Positives = 121/217 (55%), Gaps = 12/217 (5%)
Query 70 QIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLVLV 129
++R F+RKV I+A Q+L T +AA V + F L + + ++ L+++
Sbjct 33 ELRWGFIRKVYSIIAFQLLATVAVAATVVTVRPIALFFATT-GLGLALYIVIIITPLIVL 91
Query 130 C-VPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLF--- 185
C + +K P N++L+ + T + ++ A ++ L ++ T VVV+ L L+
Sbjct 92 CPLYYYHQKHPVNYLLLGIFTLALAFVVGLTCAFTNGKVILESVILTSVVVLSLTLYTFW 151
Query 186 ACQTKYDFTGWGTYLFVAVLCLMIFGILCIIFS-SKVVHLVYSGLATVLFCMILVYDTQQ 244
A + YDF G +LF A+ L+ F ++ I+F +V ++Y L +++FC +VYDT
Sbjct 152 AARKGYDFNFLGPFLFGALTVLIFFALIQILFPLGRVSVMIYGCLVSIIFCGYIVYDTDN 211
Query 245 VVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
++ +R+ Y D+YI+AA++LY+DII +F+ +L +
Sbjct 212 LI----KRHTY--DEYIWAAVSLYLDIINLFLYLLTV 242
> Hs22059607
Length=285
Score = 77.4 bits (189), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 64/209 (30%), Positives = 115/209 (55%), Gaps = 15/209 (7%)
Query 74 AFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLVLVCVPD 133
F+RKV IL++Q+L T + VF + ++R F+ ++ L ++ A+ +L L L+
Sbjct 80 TFLRKVYSILSLQVLLTTVTSTVFLYFESVRTFVHESPALILLFALGSLGLIFALI---- 135
Query 134 LAR-KVPTNFILMSLITACYSVLISCAAAASSYSSFLI--AIGATFVVVVGLMLFACQTK 190
L R K P N L+ T + ++ A + Y ++I A T V GL ++ Q+K
Sbjct 136 LNRHKYPLNLYLLFGFTLLEA--LTVAVVVTFYDVYIILQAFILTTTVFFGLTVYTLQSK 193
Query 191 YDFTGWGTYLFVAVLCLMIFGILCIIFSSKVVHLVYSGLATVLFCMILVYDTQQVVGGKH 250
DF+ +G LF + L + G L F S+++ LV + +LFC ++YDT ++
Sbjct 194 KDFSKFGAGLFALLWILCLSGFLKFFFYSEIMELVLAAAGALLFCGFIIYDTHSLM---- 249
Query 251 RRYQYSIDDYIFAALTLYMDIIIIFMNIL 279
++ S ++Y+ AA++LY+DII +F+++L
Sbjct 250 --HKLSPEEYVLAAISLYLDIINLFLHLL 276
> CE25277
Length=133
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 50/142 (35%), Positives = 77/142 (54%), Gaps = 17/142 (11%)
Query 148 ITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCL 207
ITA YS A + S LI G + ++LFA TK D T F+ +CL
Sbjct 5 ITATYS-------ADAVLISLLITTGCS----ASIILFAATTKKDLTSCLGVAFILGICL 53
Query 208 MIFG----ILCIIFSSKVVHLVYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFA 263
M+FG I CI + + +++VY+ L +L L D Q ++GG RR + S ++YIFA
Sbjct 54 MLFGLMACIFCIFLNWQFLYIVYAVLGALLCMFYLAIDIQLIMGG--RRVEISPEEYIFA 111
Query 264 ALTLYMDIIIIFMNILAIADNS 285
A +++DI+ +F+NIL + N+
Sbjct 112 ATHVFVDILGMFLNILGVVGNA 133
> Hs22056936
Length=349
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 59/207 (28%), Positives = 94/207 (45%), Gaps = 33/207 (15%)
Query 70 QIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQN---YWLAIVAAVCALVLQL 126
++R FVRKV IL IQ+L T + A+F F +++++ N YW A A A L L
Sbjct 101 KVRRVFVRKVYTILLIQLLVTLAVVALFTFCDPVKDYVQANPGWYW-ASYAVFFATYLTL 159
Query 127 VLVCVPDLA-------------------------RKVPTNFILMSLITACYSVLISCAAA 161
P A R P N IL+++ T + L ++
Sbjct 160 ACCSGPRYANGLRRNLWEYEQDHENSGPSPASHQRHFPWNLILLTVFTLSMAYLTGMLSS 219
Query 162 ASSYSSFLIAIGATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLMIFG-ILCIIFSSK 220
+ +S L+ +G T +V + + +F+ QTK+DFT LFV ++ L G IL I+ +
Sbjct 220 YYNTTSVLLCLGITALVCLSVTVFSFQTKFDFTSCQGVLFVLLMTLFFSGLILAILLPFQ 279
Query 221 VV---HLVYSGLATVLFCMILVYDTQQ 244
V H VY+ L +F + D ++
Sbjct 280 YVPWLHAVYAALGAGVFTLPSSLDPRE 306
> CE02441
Length=184
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 47/157 (29%), Positives = 79/157 (50%), Gaps = 17/157 (10%)
Query 130 CVPDLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGA---TFVVVVGLMLFA 186
C P L ++ P+ I C S C + + + A T +VV L +
Sbjct 34 CAPRLRQRSPSQLCSFGRIHRCSS----CHYGMLTLFEAKVVLEAAVITGLVVASLFAYT 89
Query 187 CQTKYDFT-GWGTYLFVAVLCLMIF-GILCIIFSSKVVHLVYSGLATVLFCMILVYDTQQ 244
Q K DF+ G+ + ++LC++++ GI + F S V+ V + LFC++LV D
Sbjct 90 LQNKRDFSVGYAS--MGSLLCVLLWAGIFQMFFMSPAVNFVINVFGAGLFCVLLVIDLDM 147
Query 245 VVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
++ Y++S +DYI A ++LYMDI+ +F+ IL I
Sbjct 148 IM------YRFSPEDYICACVSLYMDILNLFIRILQI 178
> YNL305c
Length=297
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 65/257 (25%), Positives = 109/257 (42%), Gaps = 51/257 (19%)
Query 71 IRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQLVLVC 130
IR F+ KV +L+ Q+L + +L+NF++ + L + V +LV + L
Sbjct 46 IRQRFMHKVYSLLSCQLLASLSFCYWASVSTSLQNFIMSHIALFYICMVVSLVSCIWLAV 105
Query 131 VP---DLARKVPTNFI---------------------------LMSLITACYSVLISCAA 160
P D VP + L+S+ T + +S
Sbjct 106 SPRPEDYEASVPEPLLTGSSEEPAQEQRRLPWYVLSSYKQKLTLLSIFTLSEAYCLSLVT 165
Query 161 AASSYSSFLIAIGATFVVVVGLMLFACQTKYD-----------FTGWGTYLFVAV-LCLM 208
A + L A+ T +VVVG+ L A +++ + WG ++ + + L +
Sbjct 166 LAYDKDTVLSALLITTIVVVGVSLTALSERFENVLNSATSIYYWLNWGLWIMIGMGLTAL 225
Query 209 IFGILCIIFSSKVVHLVYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLY 268
+FG SSK +L+Y L +LF L DTQ + R Y D+ + A+ LY
Sbjct 226 LFG--WNTHSSK-FNLLYGWLGAILFTAYLFIDTQLIF-----RKVYP-DEEVRCAMMLY 276
Query 269 MDIIIIFMNILAIADNS 285
+DI+ +F++IL I NS
Sbjct 277 LDIVNLFLSILRILANS 293
> 7292595
Length=341
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 39/59 (66%), Gaps = 4/59 (6%)
Query 225 VYSGLATVLFCMILVYDTQQVVGGK--HRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
+Y GL VLF L+YDTQ++V + +Y Y+ D I A++++YMD++ IF+ I+ I
Sbjct 277 LYGGL--VLFSGFLLYDTQRMVRRAEVYPQYSYTPYDPINASMSIYMDVLNIFIRIVTI 333
> CE02442
Length=130
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 20/41 (48%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 71 IRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNY 111
IR AF+RKVLGI+ Q+LFT GI A +P L + Y
Sbjct 69 IRIAFLRKVLGIVGFQLLFTIGICAAIYNIPNSNQLLQKQY 109
> At4g14730
Length=100
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 57 NGNGLQISEDISVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNY 111
GN L S ++R AF+RK+ IL++Q+L T G++AV FV + F+ + +
Sbjct 10 GGNELYPGMKESSELRWAFIRKLYSILSLQLLVTVGVSAVVYFVRPIPEFITETH 64
> CE12160
Length=342
Score = 37.7 bits (86), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 25/68 (36%), Positives = 37/68 (54%), Gaps = 12/68 (17%)
Query 224 LVYSGLATVLFCMILVYDTQQVV----GGKHRRYQYSID------DYIFAALTLYMDIII 273
+VY GL +LF L+YDTQ++V H Y D D I A +++YMD++
Sbjct 267 VVYGGL--ILFSAFLLYDTQRLVKKAENHPHSSQLYGSDMQIRSFDPINAQMSIYMDVLN 324
Query 274 IFMNILAI 281
IFM ++ I
Sbjct 325 IFMRLVMI 332
> 7298804
Length=305
Score = 36.2 bits (82), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 32/56 (57%), Gaps = 4/56 (7%)
Query 225 VYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDY--IFAALTLYMDIIIIFMNI 278
+Y GL +LF L+YDTQ++V QYS Y I AL +YMD + IF+ I
Sbjct 240 LYGGL--ILFSGFLLYDTQRIVKSAELYPQYSKFPYDPINHALAIYMDALNIFIRI 293
> At4g15470_1
Length=331
Score = 34.7 bits (78), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 28/48 (58%), Gaps = 3/48 (6%)
Query 60 GLQISEDISVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFL 107
GL E+ Q+R F+RKV GIL+ Q+L T I+AV P + + L
Sbjct 37 GLSYGEN---QLRWGFIRKVYGILSAQLLLTTLISAVVVLNPPVNDLL 81
> 7295121
Length=245
Score = 34.3 bits (77), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 46/186 (24%), Positives = 79/186 (42%), Gaps = 18/186 (9%)
Query 103 LRNFLLQNYWLAIVAAVCALVLQLVLVCVPDLARKVPTNFILMSLITACYSVLIS----- 157
+R+FL L ++AAV LVL L L D + T ++ C +
Sbjct 53 MRDFL----DLGVLAAVATLVLVLGLHFYKDDGKNYYTRLGMLYAFGFCSGQTLGPLLGY 108
Query 158 -CAAAASSYSSFLIAIGATFVVVVGLMLFACQTKYDFTGWGTYLFVAVLCLM-IFGILCI 215
C+ + S L TF+ + L A Q KY + G + V+V+ M + + +
Sbjct 109 ICSINPAIILSALTGTFVTFISLSLSALLAEQGKYLYLGG---MLVSVINTMALLSLFNM 165
Query 216 IFSSKVVHLVYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIF 275
+F S V + + + +VYDTQ +V + + D + AL L+ D++ +F
Sbjct 166 VFKSYFVQVTQLYVGVFVMAAFIVYDTQNIV----EKCRNGNRDVVQHALDLFFDVLSMF 221
Query 276 MNILAI 281
+L I
Sbjct 222 RRLLII 227
> Hs5453924
Length=469
Score = 33.9 bits (76), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 51/117 (43%), Gaps = 16/117 (13%)
Query 15 QQLFRSGYGLAQLRYLQHEDK-----------QADLSLLIE-SAQHKQVSSPPPNGNGLQ 62
QQ + SG G+ +L LQ E+K S+L E S +H + PP
Sbjct 70 QQHWGSGVGVKKLCELQPEEKCCVVGTLFKAMPLQPSILREVSEEHNLLPQPP---RSKY 126
Query 63 ISEDISVQIRHAFVR-KVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAA 118
I D + + R K+ G + + L T + AVFG V FL+++Y A +A
Sbjct 127 IHPDDELVLEDELQRIKLKGTIDVSKLVTGTVLAVFGSVRDDGKFLVEDYCFADLAP 183
> 7297457
Length=242
Score = 33.1 bits (74), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 45/169 (26%), Positives = 78/169 (46%), Gaps = 30/169 (17%)
Query 133 DLARKVPTNFILMSLITACYSVLISCAAAASSYSSFLIAIGATFVVVVGLMLFACQTKYD 192
L RKVP N+I+++ I ++ I A + L+ A +VVV LML +
Sbjct 82 KLGRKVPFNYIIIAAIVESSTIYI--AMEQKHNENRLVNFYAG-IVVVALMLASI----- 133
Query 193 FTGWGTYLFVAVL-------CLM-IFGILCIIFSSKVVHLVYSGL--------ATVLFCM 236
WG Y + ++ CL+ I I+ IIF V+ + Y G+ A V CM
Sbjct 134 --FWGAYFPMFIVPGDLLLSCLVAIANIMMIIFFINVLFINYQGIYFAVRNTFALVAVCM 191
Query 237 ILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAIADNS 285
++ T ++ R++ ++Y+F ++ + +I+ ILAI+ S
Sbjct 192 VMY--TATII--HDRQFDVPKNEYLFLSVLQFFSYMILHERILAISFTS 236
> 7298040
Length=529
Score = 32.7 bits (73), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 46/97 (47%), Gaps = 10/97 (10%)
Query 66 DISVQIRHAFVRKVLGILAIQILFTFGIAAVFGFVPTLRNFLLQNYWLAIVAAVCALVLQ 125
D V++R + + GI A+ I+ G+ + G R +LL +VA C V+
Sbjct 7 DGVVRLRLKVAQWIYGIAALFIVLAIGLLILPGL---FRRYLLVP---DVVATYCFFVIG 60
Query 126 LVLVCV----PDLARKVPTNFILMSLITACYSVLISC 158
LV +CV L RK P N+I+ I AC ++ C
Sbjct 61 LVTLCVYVNVTWLRRKFPFNWIVSCCIAACLALGTVC 97
> Hs14757108
Length=237
Score = 31.6 bits (70), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 33/57 (57%), Gaps = 6/57 (10%)
Query 225 VYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
+Y GL V+ C +++DTQ ++ + ++ DYI+ + L++D I +F ++ I
Sbjct 174 LYVGL--VVMCGFVLFDTQLII----EKAEHGDQDYIWHCIDLFLDFITVFRKLMMI 224
> 7304129
Length=117
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 48/90 (53%), Gaps = 6/90 (6%)
Query 183 MLFACQTKYDFTGWGTYLFVAVLCLMIFGI-LCIIFSSKVVHLVYSGLATVLFCMILVYD 241
ML++ Q ++ FT ++ +IFG+ L + + + V+S +A + ++YD
Sbjct 1 MLYSSQKRFRFTQIRGISIIS----LIFGLFLLLAYRLNRMMEVFSAMACTVEAWYIIYD 56
Query 242 TQQVVGGKHRRYQYSIDDYIFAALTLYMDI 271
T ++ G+H Y +++++AA ++ D+
Sbjct 57 THYMLCGRH-GYNIGPEEFVYAACNIHCDL 85
> 7295330
Length=741
Score = 30.4 bits (67), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 30/63 (47%), Gaps = 10/63 (15%)
Query 71 IRHAFVRKVLGI-LAIQILFTFGIAAVFGFV---------PTLRNFLLQNYWLAIVAAVC 120
+RHA RK+ AI +L FG+ FG + P L + WLAIVA V
Sbjct 598 MRHAMPRKLTATGQAIAVLAFFGLGKGFGALIGLARDERDPKLEFWSCTYQWLAIVACVV 657
Query 121 ALV 123
AL+
Sbjct 658 ALI 660
> CE16334
Length=581
Score = 30.0 bits (66), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 16/63 (25%), Positives = 29/63 (46%), Gaps = 12/63 (19%)
Query 192 DFTGWGTYLFVAVLCLMIFGILCIIFSSKVVHLVYSGLATVLFCMILVYDTQQVVGGKHR 251
D W Y V C +IFG C I +G+ +V C++++Y+++ +V +
Sbjct 439 DHRHWKIY--VPYFCSLIFGAWCSI----------TGVTSVFTCLLMIYESRNIVSKETL 486
Query 252 RYQ 254
Q
Sbjct 487 NMQ 489
> Hs4507433
Length=237
Score = 29.6 bits (65), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 32/57 (56%), Gaps = 6/57 (10%)
Query 225 VYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
+Y GL V+ C ++ DTQ ++ + ++ DYI+ + L++D I +F ++ I
Sbjct 174 LYVGL--VVMCGFVLVDTQLII----EKAEHGDQDYIWHCIDLFLDFITVFRKLMMI 224
> At5g47120
Length=247
Score = 29.6 bits (65), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 32/57 (56%), Gaps = 6/57 (10%)
Query 225 VYSGLATVLFCMILVYDTQQVVGGKHRRYQYSIDDYIFAALTLYMDIIIIFMNILAI 281
+Y GL ++F +V DTQ+++ H DY+ +LTL+ D + +F+ IL I
Sbjct 180 LYFGL--LIFVGYMVVDTQEIIEKAHLGDM----DYVKHSLTLFTDFVAVFVRILII 230
> CE26205
Length=1641
Score = 29.3 bits (64), Expect = 9.1, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 7 LKEAYDAQQQLFRSGYGLAQLRYLQHEDKQADLSLL-IESAQHKQVSSPPPNG 58
+K+A Q + ++GY LA + LQHE +QA L ++ KQ S NG
Sbjct 502 VKQAEPLQAEPPQNGYRLAAAQALQHEPRQASEPLQNVQPEPSKQASELVQNG 554
Lambda K H
0.331 0.143 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6304412188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40