bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0059_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
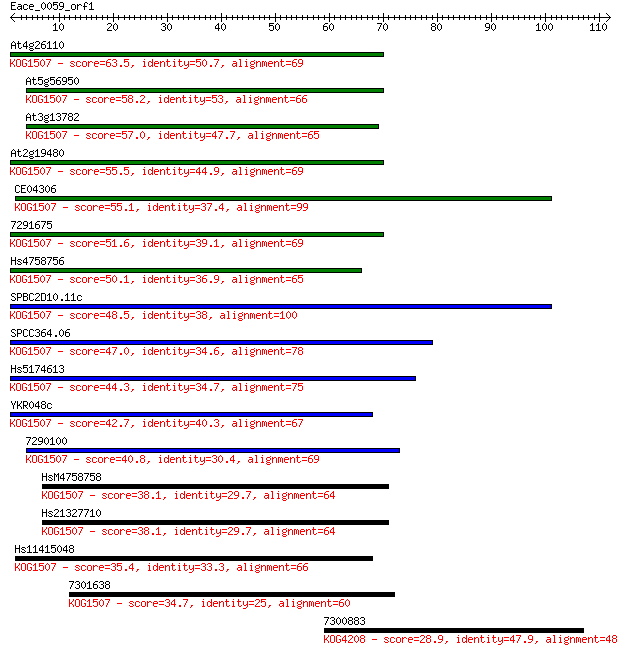
At4g26110 63.5 9e-11
At5g56950 58.2 3e-09
At3g13782 57.0 8e-09
At2g19480 55.5 2e-08
CE04306 55.1 3e-08
7291675 51.6 3e-07
Hs4758756 50.1 1e-06
SPBC2D10.11c 48.5 3e-06
SPCC364.06 47.0 8e-06
Hs5174613 44.3 5e-05
YKR048c 42.7 1e-04
7290100 40.8 6e-04
HsM4758758 38.1 0.004
Hs21327710 38.1 0.005
Hs11415048 35.4 0.025
7301638 34.7 0.040
7300883 28.9 2.7
> At4g26110
Length=372
Score = 63.5 bits (153), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 49/69 (71%), Gaps = 2/69 (2%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPR 60
T+ IT+ D SFFN F+ E+P +D + +DE+ E+LQ ++E DYDIG TIR+KIIPR
Sbjct 233 TKPITKLEDCESFFNFFSPPEVPDED--EDIDEERAEDLQNLMEQDYDIGSTIREKIIPR 290
Query 61 AVDWYLGQV 69
AV W+ G+
Sbjct 291 AVSWFTGEA 299
> At5g56950
Length=368
Score = 58.2 bits (139), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 35/66 (53%), Positives = 46/66 (69%), Gaps = 2/66 (3%)
Query 4 ITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVD 63
IT+T D SFFN F ++P DD + +DE+ EELQ ++E DYDIG TIR+KIIP AV
Sbjct 235 ITKTEDCESFFNFFNPPQVPDDD--EDIDEERAEELQNLMEQDYDIGSTIREKIIPHAVS 292
Query 64 WYLGQV 69
W+ G+
Sbjct 293 WFTGEA 298
> At3g13782
Length=329
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 31/68 (45%), Positives = 45/68 (66%), Gaps = 3/68 (4%)
Query 4 ITETVDANSFFNLFADHEIPSDDKLQQMDEKEV---EELQMVVEADYDIGITIRDKIIPR 60
+T+T + SFFN F EIP D++ D+ + EELQ +++ DYDI +TIRDK+IP
Sbjct 241 MTKTENCESFFNFFKPPEIPEIDEVDDYDDFDTIMTEELQNLMDQDYDIAVTIRDKLIPH 300
Query 61 AVDWYLGQ 68
AV W+ G+
Sbjct 301 AVSWFTGE 308
> At2g19480
Length=379
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 43/69 (62%), Gaps = 2/69 (2%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPR 60
T+ IT+T D SFFN F+ ++P DD+ D + + QM E DYDIG TI++KII
Sbjct 232 TKPITKTEDCESFFNFFSPPQVPDDDEDLDDDMADELQGQM--EHDYDIGSTIKEKIISH 289
Query 61 AVDWYLGQV 69
AV W+ G+
Sbjct 290 AVSWFTGEA 298
> CE04306
Length=316
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/99 (37%), Positives = 58/99 (58%), Gaps = 12/99 (12%)
Query 2 RTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRA 61
+ +T+TV A+SFFN F P K ++ ++++ E+ + +E DY++G IRD IIPRA
Sbjct 229 KFLTKTVKADSFFNFFE----PPKSKDERNEDEDDEQAEEFLELDYEMGQAIRDTIIPRA 284
Query 62 VDWYLGQVDDDSDFDDYDANDEEFDSEESDEEEDSSDEE 100
V +Y G++ D FD F E+ D+ D SD+E
Sbjct 285 VLFYTGELQSDDMFD--------FPGEDGDDVSDFSDDE 315
> 7291675
Length=370
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 44/70 (62%), Gaps = 8/70 (11%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEE-LQMVVEADYDIGITIRDKIIP 59
RTI + V +SFFN F+ E+PSD ++EV++ Q ++ D++IG +R +IIP
Sbjct 261 VRTIVKQVPTDSFFNFFSPPEVPSD-------QEEVDDDSQQILATDFEIGHFLRARIIP 313
Query 60 RAVDWYLGQV 69
+AV +Y G +
Sbjct 314 KAVLYYTGDI 323
> Hs4758756
Length=391
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 41/65 (63%), Gaps = 7/65 (10%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPR 60
RT+T+TV +SFFN FA E+P L ++ + ++ AD++IG +R++IIPR
Sbjct 285 VRTVTKTVSNDSFFNFFAPPEVPESGDLD-------DDAEAILAADFEIGHFLRERIIPR 337
Query 61 AVDWY 65
+V ++
Sbjct 338 SVLYF 342
> SPBC2D10.11c
Length=379
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 61/102 (59%), Gaps = 12/102 (11%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPR 60
TR + TV +SFFN F+ ++ D+ +D+K ++E DY +G +D+IIP
Sbjct 279 TRLVRTTVPNDSFFNFFSPPQLDDDESDDGLDDK-----TELLELDYQLGEVFKDQIIPL 333
Query 61 AVDWYLGQVDDDSDFDDYDANDEEFDSEE--SDEEEDSSDEE 100
A+D +L ++ D D++ DEE DSE+ +DEE+ SSD+E
Sbjct 334 AIDCFL----EEGDLSDFNQMDEE-DSEDAYTDEEDLSSDDE 370
> SPCC364.06
Length=393
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 44/78 (56%), Gaps = 4/78 (5%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPR 60
TR + +V +SFFN F P ++ +E E EL ++E DY IG ++K+IPR
Sbjct 274 TRVVKVSVPRDSFFNFFN----PPTPPSEEDEESESPELDELLELDYQIGEDFKEKLIPR 329
Query 61 AVDWYLGQVDDDSDFDDY 78
AV+W+ G+ ++D +
Sbjct 330 AVEWFTGEALALENYDGF 347
> Hs5174613
Length=375
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 45/76 (59%), Gaps = 7/76 (9%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPR 60
RTIT+ V SFFN F + D + +DE + + + +D++IG R++I+PR
Sbjct 277 VRTITKQVPNESFFNFFNPLKASGDG--ESLDE----DSEFTLASDFEIGHFFRERIVPR 330
Query 61 AVDWYLGQ-VDDDSDF 75
AV ++ G+ ++DD +F
Sbjct 331 AVLYFTGEAIEDDDNF 346
> YKR048c
Length=417
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/67 (40%), Positives = 41/67 (61%), Gaps = 4/67 (5%)
Query 1 TRTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPR 60
RTI + SFFN F +I ++D+ ++++E E L + DY IG ++DK+IPR
Sbjct 300 VRTIEKITPIESFFNFFDPPKIQNEDQDEELEEDLEERLAL----DYSIGEQLKDKLIPR 355
Query 61 AVDWYLG 67
AVDW+ G
Sbjct 356 AVDWFTG 362
> 7290100
Length=362
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 37/69 (53%), Gaps = 7/69 (10%)
Query 4 ITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVD 63
+T+ + SFF FA P L DEK ++++ D+++G +R +I+P+AV
Sbjct 280 VTKVMPRESFFRFFAP---PQALDLSLADEKT----KLILGNDFEVGFLLRTQIVPKAVL 332
Query 64 WYLGQVDDD 72
+Y G + D
Sbjct 333 FYTGDLVDS 341
> HsM4758758
Length=506
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 36/64 (56%), Gaps = 7/64 (10%)
Query 7 TVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVDWYL 66
V SFFN F+ EIP KL+ ++ +++ D++IG + D +I +++ +Y
Sbjct 433 VVPNASFFNFFSPPEIPMIGKLEPRED-------AILDEDFEIGQILHDNVILKSIYYYT 485
Query 67 GQVD 70
G+V+
Sbjct 486 GEVN 489
> Hs21327710
Length=506
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 36/64 (56%), Gaps = 7/64 (10%)
Query 7 TVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVDWYL 66
V SFFN F+ EIP KL+ ++ +++ D++IG + D +I +++ +Y
Sbjct 433 VVPNASFFNFFSPPEIPMIGKLEPRED-------AILDEDFEIGQILHDNVILKSIYYYT 485
Query 67 GQVD 70
G+V+
Sbjct 486 GEVN 489
> Hs11415048
Length=460
Score = 35.4 bits (80), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 14/66 (21%)
Query 2 RTITETVDANSFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRA 61
RT+TE +SFFN F+ H I S+ + D + D+ +G +R IIPR+
Sbjct 359 RTVTEDFPKDSFFNFFSPHGITSNGR----DGND----------DFLLGHNLRTYIIPRS 404
Query 62 VDWYLG 67
V ++ G
Sbjct 405 VLFFSG 410
> 7301638
Length=272
Score = 34.7 bits (78), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 36/60 (60%), Gaps = 5/60 (8%)
Query 12 SFFNLFADHEIPSDDKLQQMDEKEVEELQMVVEADYDIGITIRDKIIPRAVDWYLGQVDD 71
SFF F+ +P D +D + + +++ D+++G +++++IP+AV ++ G++ D
Sbjct 190 SFFEFFSPPLLPED----TLDPNYCD-VNAMLQNDFEVGFYLKERVIPKAVIFFTGEIAD 244
> 7300883
Length=356
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 32/60 (53%), Gaps = 12/60 (20%)
Query 59 PRAVDWYLGQVDDDSDFDDY-DANDEEFDSEESDEEEDS------SDEE-----PPRPHK 106
P D +++DSD +DY DA+D+E D E +E+ DS SDEE PP+ K
Sbjct 240 PTLEDLLGNTINEDSDDEDYVDASDDESDLEAEEEQADSEQTSDDSDEEEDLSPPPKQKK 299
Lambda K H
0.310 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40