bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0067_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
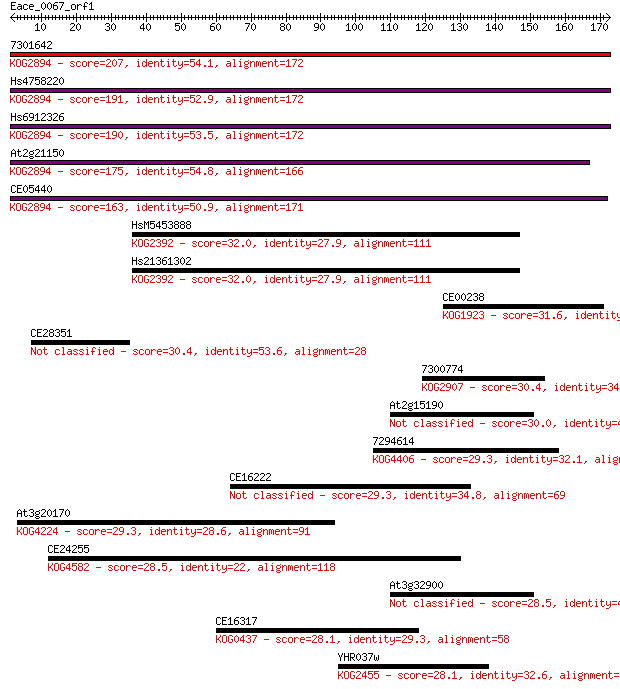
7301642 207 1e-53
Hs4758220 191 7e-49
Hs6912326 190 1e-48
At2g21150 175 5e-44
CE05440 163 1e-40
HsM5453888 32.0 0.58
Hs21361302 32.0 0.63
CE00238 31.6 0.87
CE28351 30.4 1.8
7300774 30.4 1.9
At2g15190 30.0 2.1
7294614 29.3 3.5
CE16222 29.3 4.1
At3g20170 29.3 4.1
CE24255 28.5 6.4
At3g32900 28.5 6.9
CE16317 28.1 8.9
YHR037w 28.1 9.3
> 7301642
Length=359
Score = 207 bits (526), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 93/172 (54%), Positives = 133/172 (77%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD+ER+ + R++L +E+ ++ E K +++TFSYWDGSGHRR + + +G +
Sbjct 176 DTSFLPDREREEQENRLREQLRQEWVMQQAELKDEDISITFSYWDGSGHRRNVLMKKGNS 235
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ +FL++ L KEF+EL++V + LMYVKEDLILPH+ +F++ I +KARGKSGPLF F
Sbjct 236 IYQFLQKCLELLRKEFIELKTVMADQLMYVKEDLILPHHYSFYDFIVTKARGKSGPLFQF 295
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DDVR+ +D EKE+SHAGK++ + W+ERNKHIFPASRWE ++PTK+YD
Sbjct 296 DVHDDVRMISDASVEKEESHAGKVLLRSWYERNKHIFPASRWEPYDPTKSYD 347
> Hs4758220
Length=339
Score = 191 bits (485), Expect = 7e-49, Method: Compositional matrix adjust.
Identities = 91/172 (52%), Positives = 131/172 (76%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ K+E+ K + +TFSYWDGSGHRR +++ +G T
Sbjct 163 DTSFLPDRDREEEENRLREELRQEWEAKQEKIKSEEIEITFSYWDGSGHRRTVKMRKGNT 222
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
+ +FL++A L K+F ELRS E LMY+KEDLI+PH+ +F++ I +KARGKSGPLF+F
Sbjct 223 MQQFLQKALEILRKDFSELRSAGVEQLMYIKEDLIIPHHHSFYDFIVTKARGKSGPLFNF 282
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DDVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 283 DVHDDVRLLSDATVEKDESHAGKVVLRSWYEKNKHIFPASRWEPYDPEKKWD 334
> Hs6912326
Length=325
Score = 190 bits (483), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 92/172 (53%), Positives = 130/172 (75%), Gaps = 0/172 (0%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPD++R+ + R+ L +E+ + E+ K + VTFSYWDGSGHRR +++ +G T
Sbjct 149 DTSFLPDRDREEEENRLREELRQEWEAQREKVKDEEMEVTFSYWDGSGHRRTVRVRKGNT 208
Query 61 VGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHF 120
V +FL++A + L K+F+ELRS E LM++KEDLILPH TF++ I ++ARGKSGPLF F
Sbjct 209 VQQFLKKALQGLRKDFLELRSAGVEQLMFIKEDLILPHYHTFYDFIIARARGKSGPLFSF 268
Query 121 DAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKTYD 172
D DDVR+ +D EK++SHAGK+V + W+E+NKHIFPASRWE ++P K +D
Sbjct 269 DVHDDVRLLSDATMEKDESHAGKVVLRSWYEKNKHIFPASRWEAYDPEKKWD 320
> At2g21150
Length=383
Score = 175 bits (443), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 91/185 (49%), Positives = 129/185 (69%), Gaps = 19/185 (10%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMP-----LTVTFSYWDGSGHRRRIQ- 54
+T++LPD ER+A+ AER+RL +++ R++E+ K P L +T+SYWDG+GHRR IQ
Sbjct 159 ETNFLPDSEREAEEQAERERLKKQWLREQEQIKSCPFSNEPLEITYSYWDGTGHRRVIQA 218
Query 55 -------------ILRGLTVGEFLERAKRELEKEFVELRSVPTESLMYVKEDLILPHNIT 101
+ +G +G FL +++L +F E+R+ E+L+YVKEDLI+PH +
Sbjct 219 STNYTFVILLEPYVRKGDPIGNFLRAVQQQLAPDFREIRTASVENLLYVKEDLIIPHQHS 278
Query 102 FFELIKSKARGKSGPLFHFDAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIFPASR 161
F+ELI +KARGKSGPLFHFD +DVR D EK++SHAGK+V++ W+E+NKHIFPASR
Sbjct 279 FYELIINKARGKSGPLFHFDVHEDVRTIADATIEKDESHAGKVVERHWYEKNKHIFPASR 338
Query 162 WEVFN 166
WE N
Sbjct 339 WETLN 343
> CE05440
Length=393
Score = 163 bits (413), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 87/213 (40%), Positives = 122/213 (57%), Gaps = 42/213 (19%)
Query 1 DTSYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLT 60
DTS+LPDKER+ L +++ L E+R K++ EK +TV ++YWDGS HR+ ++I +G T
Sbjct 171 DTSFLPDKEREEFLRKKKESLAAEWRVKQDAEKNEEITVAYAYWDGSSHRKNMKIKKGNT 230
Query 61 VGEFLERAK------------------------------------------RELEKEFVE 78
+ + L RA + L+KEF E
Sbjct 231 ISQCLGRAIEKAIYEIRPFRAKTHKLYFPSKIFFNCFFFSTNQKIFINIHFQALKKEFTE 290
Query 79 LRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGPLFHFDAFDDVRVFNDVRKEKED 138
L+S E+LM+VKEDLI+PH TF + I +KA GK+GPLF FD+ DVR+ D + +
Sbjct 291 LKSCTAENLMFVKEDLIIPHFYTFQDFIVTKAMGKTGPLFVFDSASDVRIRQDAALDYGE 350
Query 139 SHAGKIVDKKWFERNKHIFPASRWEVFNPTKTY 171
SH KIV + W+E+NKHI+PASRWE F P+K Y
Sbjct 351 SHPAKIVLRSWYEKNKHIYPASRWEPFVPSKKY 383
> HsM5453888
Length=427
Score = 32.0 bits (71), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 50/119 (42%), Gaps = 9/119 (7%)
Query 36 PLTVTFSYWD---GSGHRRRIQILRGLTVGEFLERAKRELEKEFVELR---SVPTESL-M 88
PL+++ +Y G+ R QIL GL E ++ ++ + F L ++P L
Sbjct 79 PLSISAAYAMLSLGACSHSRSQILEGLGF-NLTELSESDVHRGFQHLLHTLNLPGHGLET 137
Query 89 YVKEDLILPHNITFF-ELIKSKARGKSGPLFHFDAFDDVRVFNDVRKEKEDSHAGKIVD 146
V L L HN+ F + + LFH + +D V + + GKIVD
Sbjct 138 RVGSALFLSHNLKFLAKFLNDTMAVYEAKLFHTNFYDTVGTIQLINDHVKKETRGKIVD 196
> Hs21361302
Length=427
Score = 32.0 bits (71), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 50/119 (42%), Gaps = 9/119 (7%)
Query 36 PLTVTFSYWD---GSGHRRRIQILRGLTVGEFLERAKRELEKEFVELR---SVPTESL-M 88
PL+++ +Y G+ R QIL GL E ++ ++ + F L ++P L
Sbjct 79 PLSISAAYAMLSLGACSHSRSQILEGLGF-NLTELSESDVHRGFQHLLHTLNLPGHGLET 137
Query 89 YVKEDLILPHNITFF-ELIKSKARGKSGPLFHFDAFDDVRVFNDVRKEKEDSHAGKIVD 146
V L L HN+ F + + LFH + +D V + + GKIVD
Sbjct 138 RVGSALFLSHNLKFLAKFLNDTMAVYEAKLFHTNFYDTVGTIQLINDHVKKETRGKIVD 196
> CE00238
Length=732
Score = 31.6 bits (70), Expect = 0.87, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 22/48 (45%), Gaps = 2/48 (4%)
Query 125 DVRVFND--VRKEKEDSHAGKIVDKKWFERNKHIFPASRWEVFNPTKT 170
D+ V +D VR EKE H+G + F N RWE F KT
Sbjct 630 DLMVLDDKTVRAEKEMEHSGSNIPLSEFVENAKTISKERWEHFKSLKT 677
> CE28351
Length=823
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 7 DKERDAKLAAERQRLIEEYRRKEEEEKR 34
D+ER L ERQR + E RR EEE R
Sbjct 256 DRERQEMLEVERQRRVLEDRRNSEEEIR 283
> 7300774
Length=120
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 119 HFDAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERN 153
HF+ +D +VFN ++E E G +V++K + N
Sbjct 51 HFNTYDPSKVFNRTKRESESDADGPVVERKCPKCN 85
> At2g15190
Length=775
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 24/47 (51%), Gaps = 6/47 (12%)
Query 110 ARGKSGPLFH-FDAFDDVRV-----FNDVRKEKEDSHAGKIVDKKWF 150
A+ K P+ H F D R+ + RK K S AG IVD+KWF
Sbjct 568 AKKKKHPILHPFAKVDATRLEKLAEWKKSRKNKPLSIAGNIVDRKWF 614
> 7294614
Length=476
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 105 LIKSKARGKSGPLFHFDAFDDVRVFNDVRKEKEDSHAGKIVDKKWFERNKHIF 157
L+KS R + PL F+ ++DV F D KE+ + +++ +K E N +F
Sbjct 349 LLKSFLRDLAEPLLTFELYEDVTGFLDWPKEERSRNVTQLIREKLPEENYELF 401
> CE16222
Length=408
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 37/73 (50%), Gaps = 8/73 (10%)
Query 64 FLERAKRELEKEFVELRSVPTESLMYVKEDLILP----HNITFFELIKSKARGKSGPLFH 119
+L R EL + +E+ + SL+Y+KE +L +NITF L R KS P +
Sbjct 12 YLHRENVELVQGKLEVYEI---SLLYIKEMALLVLNVLNNITFISLFIYNRRRKSQPSSY 68
Query 120 FDAFDDVRVFNDV 132
FD + +FN +
Sbjct 69 RSEFDTL-IFNQM 80
> At3g20170
Length=475
Score = 29.3 bits (64), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 38/91 (41%), Gaps = 5/91 (5%)
Query 3 SYLPDKERDAKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLTVG 62
L E +A L AE+ L+ R + E K L + WD +G+R + ++RG
Sbjct 300 CILAVAEGNAVLIAEQ--LVRILRAGDNEAK---LAASDVLWDLAGYRHSVSVIRGSGAI 354
Query 63 EFLERAKRELEKEFVELRSVPTESLMYVKED 93
L R+ EF E S L Y + D
Sbjct 355 PLLIELLRDGSLEFRERISGAISQLSYNEND 385
> CE24255
Length=256
Score = 28.5 bits (62), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 26/119 (21%), Positives = 45/119 (37%), Gaps = 1/119 (0%)
Query 12 AKLAAERQRLIEEYRRKEEEEKRMPLTVTFSYWDGSGHRRRIQILRGLTVGEFLERAKRE 71
+ + R +IE + +P + S++ + HR +I L E L A E
Sbjct 2 SSIRYRRPPIIERRYERTPSYSTIPRDIHVSFYHNNYHRSKILYLNKFNAMEKLIEAAHE 61
Query 72 LEKEFVELRSVPTESLMYVKE-DLILPHNITFFELIKSKARGKSGPLFHFDAFDDVRVF 129
L R + + V+ D ILPH E+ K +F+F + ++F
Sbjct 62 LYPSNGVYRLFSVQVMECVQNPDHILPHLHIRLEVFLKKGTMIFQSIFYFQILRNFKLF 120
> At3g32900
Length=654
Score = 28.5 bits (62), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 6/47 (12%)
Query 110 ARGKSGPLFH-FDAFDDVRV-----FNDVRKEKEDSHAGKIVDKKWF 150
A K P+ H F D R+ + RK K S AG I+D KWF
Sbjct 404 AEKKKHPILHPFAKVDSTRLEKLAEWKKSRKNKPLSIAGNIIDTKWF 450
> CE16317
Length=1186
Score = 28.1 bits (61), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 17/61 (27%), Positives = 34/61 (55%), Gaps = 4/61 (6%)
Query 60 TVGEFLERAKREL---EKEFVELRSVPTESLMYVKEDLILPHNITFFELIKSKARGKSGP 116
++ EF R K + +K E+ VPTE+++YV ++ P T ++++ +A+ +G
Sbjct 911 SLAEFRARLKTYMTPKKKALKEIPEVPTEAVIYVAKEYP-PWQKTILDILEKQAKANNGA 969
Query 117 L 117
L
Sbjct 970 L 970
> YHR037w
Length=575
Score = 28.1 bits (61), Expect = 9.3, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 23/44 (52%), Gaps = 1/44 (2%)
Query 95 ILPHNITFFELIKSKARGKSGPLFHFDAFDD-VRVFNDVRKEKE 137
++P N + + RG GP+ H +FD V+V D +K+ E
Sbjct 380 VVPMNTSASPISGGNLRGFMGPVIHEQSFDKLVKVIEDAKKDPE 423
Lambda K H
0.319 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2562785186
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40