bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0084_orf2
Length=168
Score E
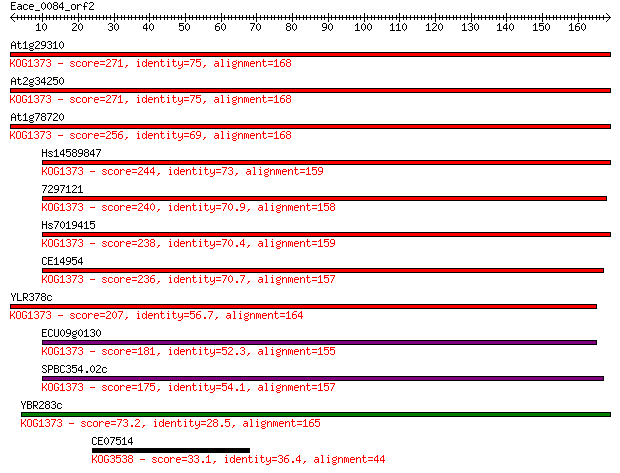
Sequences producing significant alignments: (Bits) Value
At1g29310 271 5e-73
At2g34250 271 5e-73
At1g78720 256 2e-68
Hs14589847 244 4e-65
7297121 240 1e-63
Hs7019415 238 4e-63
CE14954 236 2e-62
YLR378c 207 1e-53
ECU09g0130 181 5e-46
SPBC354.02c 175 3e-44
YBR283c 73.2 2e-13
CE07514 33.1 0.27
> At1g29310
Length=475
Score = 271 bits (692), Expect = 5e-73, Method: Compositional matrix adjust.
Identities = 126/168 (75%), Positives = 148/168 (88%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+VI LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI TG+G EFEGA++ALFH L
Sbjct 164 IVICLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINTGRGAEFEGAVIALFHML 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
TKS + AL++AFYR N PN+T+LLAT+L+FLIVIYFQGFRV L V+ + RGQQG+YP
Sbjct 224 ITKSNKVAALRQAFYRQNLPNVTNLLATVLIFLIVIYFQGFRVVLPVRSKSARGQQGSYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSN+PIILQ+ALVSNLYF+SQLLYR+F N VNLLGQW+E +
Sbjct 284 IKLFYTSNMPIILQSALVSNLYFISQLLYRKFSGNFFVNLLGQWKESE 331
> At2g34250
Length=475
Score = 271 bits (692), Expect = 5e-73, Method: Compositional matrix adjust.
Identities = 126/168 (75%), Positives = 148/168 (88%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+VI LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI TG+G EFEGA++ALFH L
Sbjct 164 IVICLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINTGRGAEFEGAVIALFHML 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
TKS + AL++AFYR N PN+T+LLAT+L+FLIVIYFQGFRV L V+ + RGQQG+YP
Sbjct 224 ITKSNKVAALRQAFYRQNLPNVTNLLATVLIFLIVIYFQGFRVVLPVRSKNARGQQGSYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSN+PIILQ+ALVSNLYF+SQLLYR+F N VNLLGQW+E +
Sbjct 284 IKLFYTSNMPIILQSALVSNLYFISQLLYRKFSGNFFVNLLGQWKESE 331
> At1g78720
Length=475
Score = 256 bits (653), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 116/168 (69%), Positives = 147/168 (87%), Gaps = 0/168 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+V+ LDELLQKGYGLGSGISLFIATNICE+I+WKAFSPTTI +G+G +FEGA++ALFH L
Sbjct 164 IVLCLDELLQKGYGLGSGISLFIATNICESIIWKAFSPTTINSGRGAQFEGAVIALFHLL 223
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
T++ + AL+EAF+R N PN+T+L AT+L+FLIVIYFQGFRV L V+ + RGQ+G+YP
Sbjct 224 ITRTDKVRALREAFFRQNLPNVTNLHATVLIFLIVIYFQGFRVVLPVRSKNARGQRGSYP 283
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
IKLFYTSN+PIILQ+ALVSN+YF+SQ+LYR+F N LVNL+G W+E +
Sbjct 284 IKLFYTSNMPIILQSALVSNIYFISQILYRKFGGNFLVNLIGTWKESE 331
> Hs14589847
Length=476
Score = 244 bits (624), Expect = 4e-65, Method: Compositional matrix adjust.
Identities = 116/159 (72%), Positives = 134/159 (84%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTTI TG+GTEFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTINTGRGTEFEGAVIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+AT+ VF +VIYFQGFRVDL +K R RGQ +YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATVFVFAVVIYFQGFRVDLPIKSARYRGQYSSYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N LVNLLGQW +V
Sbjct 290 PIILQSALVSNLYVISQMLSVRFSGNFLVNLLGQWADVS 328
> 7297121
Length=423
Score = 240 bits (612), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 112/158 (70%), Positives = 134/158 (84%), Gaps = 0/158 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG+GTEFEGA++ALFH + T++ + A
Sbjct 117 QKGYGLGSGISLFIATNICETIVWKAFSPTTVTTGRGTEFEGAVIALFHLMATRNDKVRA 176
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +LLAT+LVF +VIYFQGFRVDL +K R RGQ +YPIKLFYTSNI
Sbjct 177 LREAFYRQNLPNLMNLLATVLVFAVVIYFQGFRVDLPIKSARYRGQYSSYPIKLFYTSNI 236
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEV 167
PIILQ+ALVSNLY +SQ+L +F+ N +NLLG W +V
Sbjct 237 PIILQSALVSNLYVISQMLAVKFQGNFFINLLGVWADV 274
> Hs7019415
Length=476
Score = 238 bits (607), Expect = 4e-63, Method: Compositional matrix adjust.
Identities = 112/159 (70%), Positives = 131/159 (82%), Gaps = 0/159 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSPTT+ TG+G EFEGA++ALFH L T++ + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPTTVNTGRGMEFEGAIIALFHLLATRTDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+ATI VF +VIYFQGFRVDL +K R RGQ YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLIATIFVFAVVIYFQGFRVDLPIKSARYRGQYNTYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQEVD 168
PIILQ+ALVSNLY +SQ+L RF N+LV+LLG W +
Sbjct 290 PIILQSALVSNLYVISQMLSARFSGNLLVSLLGTWSDTS 328
> CE14954
Length=473
Score = 236 bits (602), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 111/157 (70%), Positives = 130/157 (82%), Gaps = 0/157 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIATNICETIVWKAFSP T+ TG+GTEFEGA++ALFH L T+S + A
Sbjct 170 QKGYGLGSGISLFIATNICETIVWKAFSPATMNTGRGTEFEGAVIALFHLLATRSDKVRA 229
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
L+EAFYR N PN+ +L+AT LVF +VIYFQGFRVDL +K R RGQ +YPIKLFYTSNI
Sbjct 230 LREAFYRQNLPNLMNLMATFLVFAVVIYFQGFRVDLPIKSARYRGQYSSYPIKLFYTSNI 289
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQE 166
PIILQ+ALVSNLY +SQ+L +F N +NLLG W +
Sbjct 290 PIILQSALVSNLYVISQMLAGKFGGNFFINLLGTWSD 326
> YLR378c
Length=480
Score = 207 bits (526), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 93/164 (56%), Positives = 126/164 (76%), Gaps = 0/164 (0%)
Query 1 VVIILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCL 60
+V++LDELL KGYGLGSGISLF ATNI E I W+AF+PTT+ +G+G EFEGA++A FH L
Sbjct 163 IVMLLDELLSKGYGLGSGISLFTATNIAEQIFWRAFAPTTVNSGRGKEFEGAVIAFFHLL 222
Query 61 FTKSTNMVALKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
+ AL EAFYR+N PN+ +L T+ +FL V+Y QGFR +L ++ +VRGQ G YP
Sbjct 223 AVRKDKKRALVEAFYRTNLPNMFQVLMTVAIFLFVLYLQGFRYELPIRSTKVRGQIGIYP 282
Query 121 IKLFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQW 164
IKLFYTSN PI+LQ+AL SN++ +SQ+L++++ N L+ L+G W
Sbjct 283 IKLFYTSNTPIMLQSALTSNIFLISQILFQKYPTNPLIRLIGVW 326
> ECU09g0130
Length=410
Score = 181 bits (460), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 81/155 (52%), Positives = 114/155 (73%), Gaps = 0/155 (0%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLG+G++LFIA N+CE+I+WKAFSP + TG+G EFEG+++ALFH L + A
Sbjct 107 QKGYGLGNGVNLFIAANVCESIIWKAFSPKVLFTGRGIEFEGSVIALFHLLVVRKNKFAA 166
Query 70 LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIKLFYTSNI 129
+ EAF+R N PN+ SLL+TIL+F+ VIY QG RV+L + +VRG G +PIKL YTS +
Sbjct 167 IYEAFFRQNLPNLFSLLSTILLFVFVIYLQGMRVELPTESSQVRGHVGKFPIKLLYTSTM 226
Query 130 PIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQW 164
PII Q+ +V ++ +S LY+R+ ++V +LG W
Sbjct 227 PIIAQSYIVGHISSISSFLYKRWPQYLVVRILGVW 261
> SPBC354.02c
Length=479
Score = 175 bits (444), Expect = 3e-44, Method: Compositional matrix adjust.
Identities = 85/164 (51%), Positives = 112/164 (68%), Gaps = 7/164 (4%)
Query 10 QKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNMVA 69
QKGYGLGSGISLFIAT CE I WKAFSPTT G +FEGA++ + +FT A
Sbjct 172 QKGYGLGSGISLFIATINCENIFWKAFSPTTYHIANGVQFEGAVINFVYVMFTWDNKAAA 231
Query 70 LKEAFYRS-------NAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYPIK 122
L +AF+RS PN+ + AT+LVF +VIY Q FRV++ ++ Q+ RG + +P+K
Sbjct 232 LYQAFFRSGLTSSQIQLPNLWNFFATLLVFGVVIYLQDFRVEIPIRSQKFRGYRSTFPVK 291
Query 123 LFYTSNIPIILQTALVSNLYFLSQLLYRRFKNNVLVNLLGQWQE 166
L YTSN PI+LQ+AL SNL+F S+LL+ RF +N LV LG W++
Sbjct 292 LLYTSNTPIMLQSALTSNLFFASRLLFNRFSSNFLVRFLGVWEQ 335
> YBR283c
Length=490
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 47/172 (27%), Positives = 87/172 (50%), Gaps = 7/172 (4%)
Query 4 ILDELLQKGYGLGSGISLFIATNICETIVWKAFSPTTIKTGKG--TEFEGALVALFHCLF 61
+L E++ KG+G SG + I +V F + IK G+ TE +GAL+ L L
Sbjct 166 LLAEVIDKGFGFSSGAMIINTVVIATNLVADTFGVSQIKVGEDDQTEAQGALINLIQGLR 225
Query 62 TKSTNMVA-LKEAFYRSNAPNITSLLATILVFLIVIYFQGFRVDLAVKYQRVRGQQGNYP 120
+K + + AF R PN+T+ + + + +IV Y Q RV+L ++ R RG YP
Sbjct 226 SKHKTFIGGIISAFNRDYLPNLTTTIIVLAIAIIVCYLQSVRVELPIRSTRARGTNNVYP 285
Query 121 IKLFYTSNIPIILQTALVSNLYFLS----QLLYRRFKNNVLVNLLGQWQEVD 168
IKL YT + ++ ++ ++ + QL+ + +++ ++G ++ +
Sbjct 286 IKLLYTGCLSVLFSYTILFYIHIFAFVLIQLVAKNEPTHIICKIMGHYENAN 337
> CE07514
Length=860
Score = 33.1 bits (74), Expect = 0.27, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 24 ATNICETIVWKAFSPTTIKTGKGTEFEGALVALFHCLFTKSTNM 67
AT +C + + P TI +G+G +FE A+ ++ CL + STN+
Sbjct 516 ATRVCSRLRDENAIPNTILSGEGMQFEQAMCKIW-CLISGSTNI 558
Lambda K H
0.327 0.142 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2418402922
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40