bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0091_orf1
Length=96
Score E
Sequences producing significant alignments: (Bits) Value
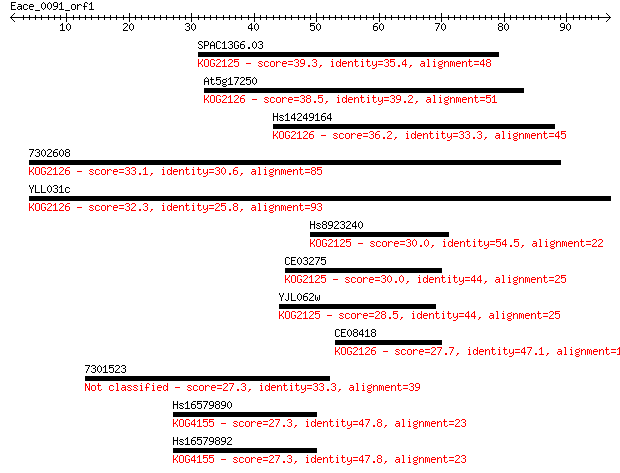
SPAC13G6.03 39.3 0.002
At5g17250 38.5 0.003
Hs14249164 36.2 0.015
7302608 33.1 0.12
YLL031c 32.3 0.20
Hs8923240 30.0 1.00
CE03275 30.0 1.2
YJL062w 28.5 3.4
CE08418 27.7 5.9
7301523 27.3 6.7
Hs16579890 27.3 7.5
Hs16579892 27.3 7.7
> SPAC13G6.03
Length=758
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 32/48 (66%), Gaps = 2/48 (4%)
Query 31 TFGILTGVTFKHLCSLLSLTLLRQHLFIWHLFAVKFLYDATAACLTYV 78
TF I++ ++ LC +S ++R HLF+W +F+ K LY+A+ A + ++
Sbjct 700 TFWIMSSISLTFLC--ISCFIMRHHLFVWSVFSPKLLYNASWASMYFL 745
> At5g17250
Length=884
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 32/51 (62%), Gaps = 2/51 (3%)
Query 32 FGILTGVTFKHLCSLLSLTLLRQHLFIWHLFAVKFLYDATAACLTYVLIVL 82
FG+++ T ++L +T+ R+HL +W LFA KF++D LT +LI L
Sbjct 830 FGVISATTVT--ATILCVTIQRRHLMVWGLFAPKFVFDVVDLILTDLLICL 878
> Hs14249164
Length=454
Score = 36.2 bits (82), Expect = 0.015, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 43 LCSLLSLTLLRQHLFIWHLFAVKFLYDATAACLTYVLIVLTLFIV 87
L L+ ++LR+HL +W +FA KF+++A ++ V ++L + +V
Sbjct 390 LACALAASILRRHLMVWKVFAPKFIFEAVGFIVSSVGLLLGIALV 434
> 7302608
Length=1077
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 43/85 (50%), Gaps = 11/85 (12%)
Query 4 SRCMLTFYQRSTAPEVVLMTTLMIGKVTFGILTGVTFKHLCSLLSLTLLRQHLFIWHLFA 63
+R L+ Y+ E V + T F +L G+ K C++L+ T+ +HL +W +FA
Sbjct 980 TRGELSLYEY----EDVFLGTGFKLATQFFMLQGL--KIFCAMLACTIHCRHLMVWKIFA 1033
Query 64 VKFLYDATAACLTYVLIVLTLFIVG 88
+F+Y+A A + L IVG
Sbjct 1034 PRFIYEALA-----TFVSLPALIVG 1053
> YLL031c
Length=1017
Score = 32.3 bits (72), Expect = 0.20, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 48/96 (50%), Gaps = 8/96 (8%)
Query 4 SRCMLTFYQRSTAPEVVLMTTLMIGKVTFG---ILTGVTFKHLCSLLSLTLLRQHLFIWH 60
S +LT + + P VL ++G++ +LT T L S + +T R+HL +W
Sbjct 919 SVALLTLWSQ---PPDVLKPQTLLGRIVSNCGILLTYNTILCLSSFIWVTHFRRHLMVWK 975
Query 61 LFAVKFLYDATAACLTYVLIVLTLFIVGIGAQRTIR 96
+F +F++ + + +T +V+T + + R I+
Sbjct 976 IFCPRFIFASLSLIVTQ--LVVTFGTIAFASGRLIK 1009
> Hs8923240
Length=604
Score = 30.0 bits (66), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 12/22 (54%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 49 LTLLRQHLFIWHLFAVKFLYDA 70
+T LR HLFIW +F+ K LY+
Sbjct 559 VTSLRYHLFIWSVFSPKLLYEG 580
> CE03275
Length=795
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 45 SLLSLTLLRQHLFIWHLFAVKFLYD 69
SL+ L + + HLFIW +++ K +YD
Sbjct 758 SLICLYIFQNHLFIWSVYSPKVVYD 782
> YJL062w
Length=830
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 44 CSLLSLTLLRQHLFIWHLFAVKFLY 68
C + + +LR HLFIW +F+ K Y
Sbjct 781 CLMTACVVLRFHLFIWSVFSPKLCY 805
> CE08418
Length=905
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 8/17 (47%), Positives = 14/17 (82%), Gaps = 0/17 (0%)
Query 53 RQHLFIWHLFAVKFLYD 69
R+HL +W +FA KF+++
Sbjct 853 RRHLMVWKIFAPKFIFE 869
> 7301523
Length=363
Score = 27.3 bits (59), Expect = 6.7, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 19/39 (48%), Gaps = 0/39 (0%)
Query 13 RSTAPEVVLMTTLMIGKVTFGILTGVTFKHLCSLLSLTL 51
R P +V+ + + + V + ILT KHLC L L
Sbjct 187 RIVVPSIVIFSVMSLCSVIYTILTNGHLKHLCGQLRENL 225
> Hs16579890
Length=1490
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 27 IGKVTFGILTGVTFKHLCSLLSL 49
+GK T+G+ VT +HL S++SL
Sbjct 875 VGKGTYGVSRAVTTQHLLSIISL 897
> Hs16579892
Length=1457
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 27 IGKVTFGILTGVTFKHLCSLLSL 49
+GK T+G+ VT +HL S++SL
Sbjct 875 VGKGTYGVSRAVTTQHLLSIISL 897
Lambda K H
0.337 0.144 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1201432980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40