bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
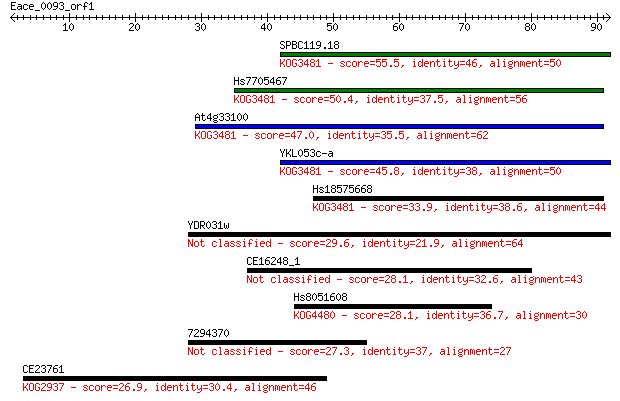
Query= Eace_0093_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
SPBC119.18 55.5 2e-08
Hs7705467 50.4 7e-07
At4g33100 47.0 1e-05
YKL053c-a 45.8 2e-05
Hs18575668 33.9 0.081
YDR031w 29.6 1.5
CE16248_1 28.1 3.6
Hs8051608 28.1 3.6
7294370 27.3 7.3
CE23761 26.9 8.6
> SPBC119.18
Length=69
Score = 55.5 bits (132), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 36/51 (70%), Gaps = 1/51 (1%)
Query 42 CSGLKEQYDRCFNHWYRRHFLRGDM-SRNCFHLFADYRLCISEELKRKGLE 91
C+ K++YD CFN WY FL+GD+ +R+C LFA+Y+ C+ + LK K ++
Sbjct 9 CTPAKKKYDACFNDWYANKFLKGDLHNRDCDELFAEYKSCLLKALKTKKID 59
> Hs7705467
Length=76
Score = 50.4 bits (119), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 34/57 (59%), Gaps = 1/57 (1%)
Query 35 MREAMDTCSGLKEQYDRCFNHWYRRHFLRGDMSRN-CFHLFADYRLCISEELKRKGL 90
M + C+ +K +YD+CFN W+ FL+GD S + C LF Y+ C+ + +K K +
Sbjct 1 MNSVGEACTDMKREYDQCFNRWFAEKFLKGDSSGDPCTDLFKRYQQCVQKAIKEKEI 57
> At4g33100
Length=92
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 1/63 (1%)
Query 29 QKRATAMREAMDTCSGLKEQYDRCFNHWYRRHFLRGDMSR-NCFHLFADYRLCISEELKR 87
+K +T+ R + C+ L+ Y CFN WY F++G + C + YR C+SE L
Sbjct 6 KKDSTSARSSTSPCADLRNAYHNCFNKWYSEKFVKGQWDKEECVAEWKKYRDCLSENLDG 65
Query 88 KGL 90
K L
Sbjct 66 KLL 68
> YKL053c-a
Length=86
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 42 CSGLKEQYDRCFNHWYRRHFLRGDMSRN-CFHLFADYRLCISEELKRKGLE 91
C+ LK +YD CFN WY FL+G N C + Y C++ L ++G++
Sbjct 13 CTDLKTKYDSCFNEWYSEKFLKGKSVENECSKQWYAYTTCVNAALVKQGIK 63
> Hs18575668
Length=91
Score = 33.9 bits (76), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 5/49 (10%)
Query 47 EQYDRCFNHWYRRHFLRGDMS-RNCFHLFADYRLCI----SEELKRKGL 90
+ YD+CF+ W+ L+GD S C +LF Y C+ +E + +GL
Sbjct 14 QSYDQCFSCWFVEKLLKGDSSGEPCSNLFKHYLQCVQKATTERISVEGL 62
> YDR031w
Length=117
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 14/64 (21%), Positives = 31/64 (48%), Gaps = 4/64 (6%)
Query 28 LQKRATAMREAMDTCSGLKEQYDRCFNHWYRRHFLRGDMSRNCFHLFADYRLCISEELKR 87
++++ +++ M CS ++YD+C R + ++ NC D R C ++K
Sbjct 49 IREKVPSVKSIMSECSEPMKKYDQCI----RDNMGTRTINENCLGFLQDLRKCAELQVKN 104
Query 88 KGLE 91
K ++
Sbjct 105 KNIK 108
> CE16248_1
Length=1259
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 22/43 (51%), Gaps = 2/43 (4%)
Query 37 EAMDTCSGLKEQYDRCFNHWYRRHFLRGDMSRNCFHLFADYRL 79
E +D +K YD + HWY ++ D S N +H + Y+L
Sbjct 430 EVLDDLKHMKHNYDTMYKHWYDMYW-DADESANEYH-YPPYKL 470
> Hs8051608
Length=316
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 44 GLKEQYDRCFNHWYRRHFLRGDMSRNCFHL 73
G KE +DR N + + FL+G++ + F L
Sbjct 40 GTKEAHDRAENTQFVKDFLKGNIKKELFKL 69
> 7294370
Length=148
Score = 27.3 bits (59), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 28 LQKRATAMREAMDTCSGLKEQYDRCFN 54
LQ T + +A+D CS ++E + C N
Sbjct 3 LQAEFTKIDDALDICSSMEEDLNECLN 29
> CE23761
Length=367
Score = 26.9 bits (58), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 27/46 (58%), Gaps = 1/46 (2%)
Query 3 HTVTDSSNSTDTQGTSSATQRTPEELQKRATAMREAMDTCSGLKEQ 48
H +SNST+++ + +T+ P +QK ++++A + S L EQ
Sbjct 36 HIFKMASNSTNSKKQNKSTEEPPSSVQKLLASLQQAQNK-SDLSEQ 80
Lambda K H
0.319 0.129 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174483934
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40