bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
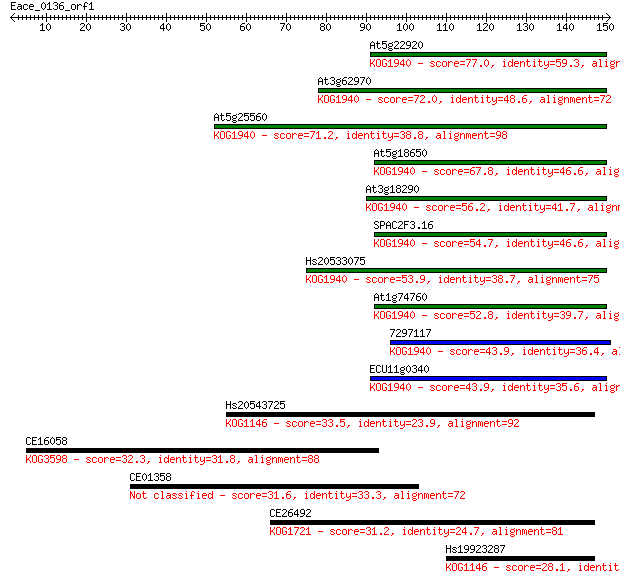
Query= Eace_0136_orf1
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
At5g22920 77.0 1e-14
At3g62970 72.0 4e-13
At5g25560 71.2 7e-13
At5g18650 67.8 8e-12
At3g18290 56.2 2e-08
SPAC2F3.16 54.7 7e-08
Hs20533075 53.9 1e-07
At1g74760 52.8 2e-07
7297117 43.9 1e-04
ECU11g0340 43.9 1e-04
Hs20543725 33.5 0.16
CE16058 32.3 0.31
CE01358 31.6 0.59
CE26492 31.2 0.67
Hs19923287 28.1 7.0
> At5g22920
Length=291
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 35/62 (56%), Positives = 46/62 (74%), Gaps = 3/62 (4%)
Query 91 QLGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYESDLVK---AHEIDRHAVEEIVCAVCET 147
GCSHYRR+CKI APCC EI+ CRHCHNEA +S ++ HE+ RH V +++C++CET
Sbjct 24 HYGCSHYRRRCKIRAPCCDEIFDCRHCHNEAKDSLHIEQHHRHELPRHEVSKVICSLCET 83
Query 148 RQ 149
Q
Sbjct 84 EQ 85
> At3g62970
Length=274
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/78 (44%), Positives = 50/78 (64%), Gaps = 6/78 (7%)
Query 78 AAAGDSAAAETAQ----QLGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYES--DLVKAHE 131
AAA DS+ Q GC HY+R+CKI APCC I+ CRHCHN++ S D + H+
Sbjct 15 AAAADSSIPRDKDFGKFQFGCEHYKRRCKIRAPCCNLIFSCRHCHNDSANSLPDPKERHD 74
Query 132 IDRHAVEEIVCAVCETRQ 149
+ R V+++VC++C+T Q
Sbjct 75 LVRQNVKQVVCSICQTEQ 92
> At5g25560
Length=272
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 56/102 (54%), Gaps = 5/102 (4%)
Query 52 ESDDGSSSSSSDSDSSADTPNPPAAEAAAGDSAAAETAQQLGCSHYRRKCKIVAPCCKEI 111
E S S + +S + T AAE+ + + GC HYRR+C I APCC EI
Sbjct 22 EMSRHSHPHSINEESESSTLERVAAESLT-NKVLDRGLMEYGCPHYRRRCCIRAPCCNEI 80
Query 112 YWCRHCHNEAYESDL----VKAHEIDRHAVEEIVCAVCETRQ 149
+ C HCH EA ++++ + H+I RH VE+++C +C T Q
Sbjct 81 FGCHHCHYEAKQNNINVDQKQRHDIPRHQVEQVICLLCGTEQ 122
> At5g18650
Length=267
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 44/60 (73%), Gaps = 2/60 (3%)
Query 92 LGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYES--DLVKAHEIDRHAVEEIVCAVCETRQ 149
GC HY+R+C+I APCC E++ CRHCHNE+ + ++ H++ R V++++C+VC+T Q
Sbjct 16 FGCKHYKRRCQIRAPCCNEVFDCRHCHNESTSTLRNIYDRHDLVRQDVKQVICSVCDTEQ 75
> At3g18290
Length=1254
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/60 (41%), Positives = 33/60 (55%), Gaps = 6/60 (10%)
Query 90 QQLGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYESDLVKAHEIDRHAVEEIVCAVCETRQ 149
Q GC HY+R CK+ A CC +++ CR CH D V H +DR V E++C C Q
Sbjct 1002 QIYGCEHYKRNCKLRAACCDQLFTCRFCH------DKVSDHSMDRKLVTEMLCMRCLKVQ 1055
> SPAC2F3.16
Length=425
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/58 (46%), Positives = 35/58 (60%), Gaps = 6/58 (10%)
Query 92 LGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYESDLVKAHEIDRHAVEEIVCAVCETRQ 149
LGCSHY R CK+ C E Y CRHCHN+A + H ++R AVE ++C +C Q
Sbjct 140 LGCSHYMRNCKVQCFDCHEWYTCRHCHNDACD------HVLERPAVENMLCMICSKVQ 191
> Hs20533075
Length=261
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 40/75 (53%), Gaps = 6/75 (8%)
Query 75 AAEAAAGDSAAAETAQQLGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYESDLVKAHEIDR 134
A A D A+ + Q GC HY R C + APCC ++Y CR CH+ + H++DR
Sbjct 1 MAATAREDGASGQERGQRGCEHYDRGCLLKAPCCDKLYTCRLCHDNN------EDHQLDR 54
Query 135 HAVEEIVCAVCETRQ 149
V+E+ C CE Q
Sbjct 55 FKVKEVQCINCEKIQ 69
> At1g74760
Length=255
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 37/58 (63%), Gaps = 6/58 (10%)
Query 92 LGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYESDLVKAHEIDRHAVEEIVCAVCETRQ 149
GC+HY+R CK++APCC +++ C CH+E E+D H +DR + +++C C Q
Sbjct 19 FGCNHYKRNCKLLAPCCDKLFTCIRCHDE--EAD----HSVDRKQITKMMCMKCLLIQ 70
> 7297117
Length=496
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 9/55 (16%)
Query 96 HYRRKCKIVAPCCKEIYWCRHCHNEAYESDLVKAHEIDRHAVEEIVCAVCETRQS 150
H ++C PCC + Y CR CH+E + H DR + E++C+ C TRQ+
Sbjct 252 HLGKQC---TPCCNKFYKCRFCHDEN------ETHHFDRKTLTELICSECNTRQT 297
> ECU11g0340
Length=254
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 6/59 (10%)
Query 91 QLGCSHYRRKCKIVAPCCKEIYWCRHCHNEAYESDLVKAHEIDRHAVEEIVCAVCETRQ 149
+ C HY C + CC +Y CR CH++A +AH +R+ V +IVC C Q
Sbjct 7 HMSCEHYSNNCLVRFECCSSLYPCRLCHDKA------EAHRANRYEVSQIVCGTCNLLQ 59
> Hs20543725
Length=1926
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 22/94 (23%), Positives = 43/94 (45%), Gaps = 2/94 (2%)
Query 55 DGSSSSSSDSDSSADTPNPPAAEAAAGDSAAAETAQQLGCSHYRRKCKIVAPCCKE-IYW 113
D S +S+ DS TP + AG + A++ + R K C E Y
Sbjct 130 DNSEGKNSNKDSGIITPEKELKVSVAGGTQPLLLAKEEDVATKRSKPTEDNKFCHEQFYQ 189
Query 114 CRHCHNEAYESDLVKAHEIDRHAVEEIV-CAVCE 146
C +C+ + + ++ H + +H+V+ ++ C +C+
Sbjct 190 CPYCNYNSRDQSRIQMHVLSQHSVQPVICCPLCQ 223
> CE16058
Length=3498
Score = 32.3 bits (72), Expect = 0.31, Method: Composition-based stats.
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 15/103 (14%)
Query 5 AEGEGVGDSQNEEEASQLTESEYS---EDDDDSEQWEEAVSSAEEGSIP--------FES 53
AE D + E +L E E + +DD S + + +SAE+ P +
Sbjct 588 AEMADANDKTDASEKQKLVEEEPTGKENEDDTSSKTAKTSTSAEKSEAPSIVDSNDKIDK 647
Query 54 DDGSSSSSSDSDSSADT----PNPPAAEAAAGDSAAAETAQQL 92
+ +SS+S+D S DT +PPAA +S T + L
Sbjct 648 EPNASSTSNDETSKDDTVPMESDPPAATEKPKESTEITTEEPL 690
> CE01358
Length=558
Score = 31.6 bits (70), Expect = 0.59, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 35/78 (44%), Gaps = 14/78 (17%)
Query 31 DDDSEQWEEAVSSAEEGSIPFESDDGSSSSSSDSDSSAD------TPNPPAAEAAAGDSA 84
DDD E V++ E+ SI +E+ DGS D D T PA+ ++ D +
Sbjct 12 DDDMNDMREMVANDEDDSIHYEAHDGSEYGGMPIDQYQDIVMFKTTDQVPASVSSRLDRS 71
Query 85 AAETAQQLGCSHYRRKCK 102
A SH+R +CK
Sbjct 72 AP--------SHWRARCK 81
> CE26492
Length=233
Score = 31.2 bits (69), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 20/92 (21%), Positives = 36/92 (39%), Gaps = 15/92 (16%)
Query 66 SSADTPNPPAAEAAAGDSAAAETAQQLGCSHYRRKCKIVAPCCKEI-----------YWC 114
S+ P P + D + A Q C R+C+ +E+ + C
Sbjct 29 SNCKKPQPLNKPSPVRDRSRTPKAPQFNC----RQCRKAFDSIRELAEHEIKNHEDQFNC 84
Query 115 RHCHNEAYESDLVKAHEIDRHAVEEIVCAVCE 146
HC ++ + + AH RH + ++CA C+
Sbjct 85 YHCDKKSESIERLAAHTAHRHGPKPVICAYCQ 116
> Hs19923287
Length=3703
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 8/38 (21%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 110 EIYWCRHCHNEAYESDLVKAHEIDRHAVEEIV-CAVCE 146
++Y C +C + + ++ H + +H+V+ ++ C +C+
Sbjct 1221 QMYQCPYCKYSNADVNRLRVHAMTQHSVQPMLRCPLCQ 1258
Lambda K H
0.304 0.119 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1852391706
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40