bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0161_orf1
Length=199
Score E
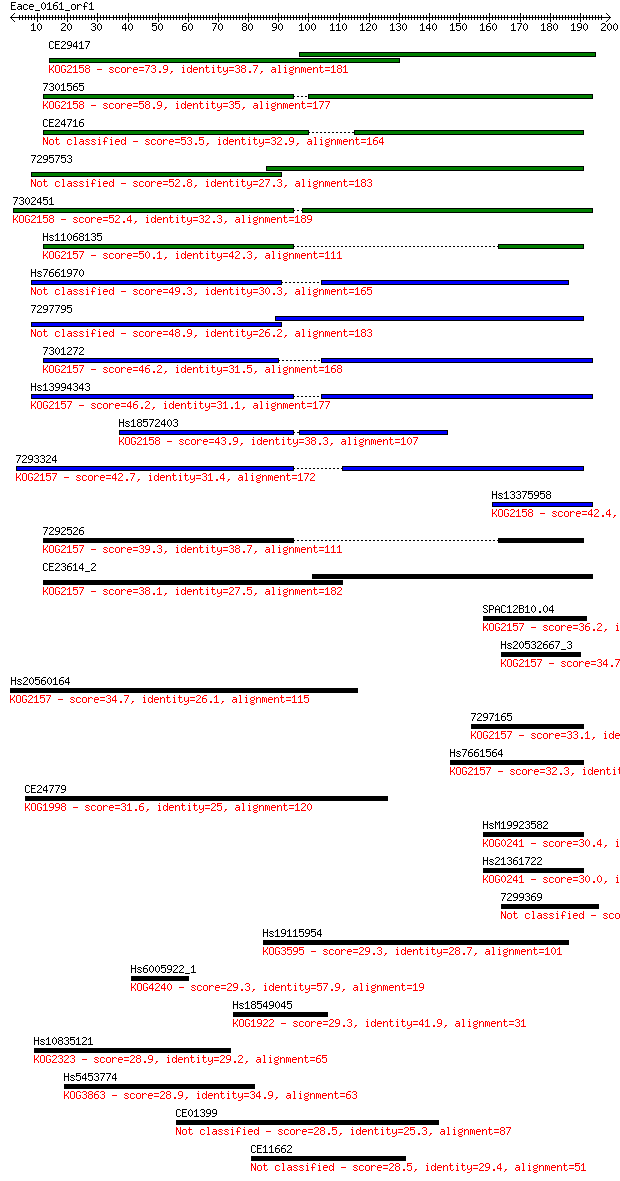
Sequences producing significant alignments: (Bits) Value
CE29417 73.9 2e-13
7301565 58.9 6e-09
CE24716 53.5 2e-07
7295753 52.8 5e-07
7302451 52.4 6e-07
Hs11068135 50.1 3e-06
Hs7661970 49.3 5e-06
7297795 48.9 7e-06
7301272 46.2 4e-05
Hs13994343 46.2 5e-05
Hs18572403 43.9 2e-04
7293324 42.7 4e-04
Hs13375958 42.4 6e-04
7292526 39.3 0.005
CE23614_2 38.1 0.011
SPAC12B10.04 36.2 0.039
Hs20532667_3 34.7 0.13
Hs20560164 34.7 0.15
7297165 33.1 0.42
Hs7661564 32.3 0.58
CE24779 31.6 1.2
HsM19923582 30.4 2.7
Hs21361722 30.0 2.8
7299369 29.3 4.9
Hs19115954 29.3 4.9
Hs6005922_1 29.3 5.7
Hs18549045 29.3 6.0
Hs10835121 28.9 6.3
Hs5453774 28.9 7.9
CE01399 28.5 8.4
CE11662 28.5 9.1
> CE29417
Length=535
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 53/98 (54%), Gaps = 7/98 (7%)
Query 97 TSNSKRLLSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVYQGEQQQQQ 156
T SKRLLS VF L++ GV ++W IK I KT + + P + L Y H + Q
Sbjct 190 TRGSKRLLSTVFHQLESRGVKTKRLWHDIKLILVKTTLAMLPEIMLHYEHHFYDSTGPQ- 248
Query 157 QQQRSRCFQFLGLDILLDADDKCWLLEVNSSPSLRVDY 194
CFQ +G D+++ D LLEVN++PSL D+
Sbjct 249 ------CFQIMGFDVMIREDGTPILLEVNAAPSLTADH 280
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 63/124 (50%), Gaps = 9/124 (7%)
Query 14 VNRFPGMWDMTKKRMLSKVLHLYQDLFPHLYDFSPPCWSFPEDRARIEEILQSS----KS 69
VN+FPGM ++ KK L+ + Q LFP Y F P W P A + + K+
Sbjct 12 VNKFPGMTELAKKISLTHSISSMQKLFPDEYAFYPNSWFLPAHLADFHAFYRKAQALGKT 71
Query 70 ET-YIIKPDGGAMGSGVQLVSRLKDIHNTSNSKRLLSDVFKD--LKAEGVDVD-KVWKSI 125
E +I+KPD GA G+G+ L++ I N + ++L+ + D L + + D +V+ I
Sbjct 72 EMWFIVKPDEGAQGTGIYLINSPNQIRNV-DQRQLVQEYVADPLLMNDKLKFDFRVYGVI 130
Query 126 KEIT 129
K I
Sbjct 131 KSIN 134
> 7301565
Length=720
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 44/83 (53%), Gaps = 1/83 (1%)
Query 12 QIVNRFPGMWDMTKKRMLSKVLHLYQDLFPHLYDFSPPCWSFPEDRARIEEILQSSKSET 71
Q +N FPGM ++ +K +LS+ L+ +FP Y P W P D + K T
Sbjct 197 QQINHFPGMIEICRKDLLSRNLNRMLKIFPQDYKIFPKTWMLPADYGDAMNYALNHK-RT 255
Query 72 YIIKPDGGAMGSGVQLVSRLKDI 94
+I+KPD GA G G+ L + LK I
Sbjct 256 FILKPDSGAQGRGIWLTNDLKTI 278
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/94 (32%), Positives = 49/94 (52%), Gaps = 6/94 (6%)
Query 100 SKRLLSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVYQGEQQQQQQQQ 159
SKR LS + ++ DV++ W ++ ++ KT + P L+ Y + G + Q
Sbjct 369 SKRKLSAINNWMRRHNYDVEEFWSNVDDVIIKTVLSAWPVLKHNYHACFPGHDKIQA--- 425
Query 160 RSRCFQFLGLDILLDADDKCWLLEVNSSPSLRVD 193
CF+ LG DIL+D K ++LEVN SPS +
Sbjct 426 ---CFEILGFDILVDWKLKPYILEVNHSPSFHTN 456
> CE24716
Length=680
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 42/76 (55%), Gaps = 9/76 (11%)
Query 115 GVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVYQGEQQQQQQQQRSRCFQFLGLDILLD 174
GVD +K+ + I+E+ K F+ + + E + +Q C++ G+DI+LD
Sbjct 455 GVDREKIQREIEEVIIKAFISTEKPIR---------EHMSRFLEQEFICYELFGIDIILD 505
Query 175 ADDKCWLLEVNSSPSL 190
D K WLLEVN SPSL
Sbjct 506 EDYKPWLLEVNISPSL 521
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 43/88 (48%), Gaps = 0/88 (0%)
Query 12 QIVNRFPGMWDMTKKRMLSKVLHLYQDLFPHLYDFSPPCWSFPEDRARIEEILQSSKSET 71
Q VN FPG + + +K L + Q+ F +D P + P DR + + L++ S
Sbjct 270 QKVNHFPGAFHIGRKDRLWMHIRKQQERFEGEFDIMPFTYILPTDRQELLKYLETDASRH 329
Query 72 YIIKPDGGAMGSGVQLVSRLKDIHNTSN 99
I+KP A G+G+ + + KD T+
Sbjct 330 VIVKPPASARGTGISVTRKPKDFPTTAT 357
> 7295753
Length=989
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 50/105 (47%), Gaps = 9/105 (8%)
Query 86 QLVSRLKDIHNTSNSKRLLSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYR 145
Q ++ +D + K L ++ L+ GV+ ++W +++ + K V + L YR
Sbjct 670 QNYAKNEDFNACQGHKWTLQSLWSCLENRGVNTKRLWATLRNLVIKGIVSGESGLNRMYR 729
Query 146 HVYQGEQQQQQQQQRSRCFQFLGLDILLDADDKCWLLEVNSSPSL 190
Q R CF+ G D+LLD + WLLE+N SPSL
Sbjct 730 ---------QNVNFRYNCFELFGFDVLLDENLVPWLLEINISPSL 765
Score = 33.1 bits (74), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 20/86 (23%), Positives = 42/86 (48%), Gaps = 3/86 (3%)
Query 8 VRSAQIVNRFPGMWDMTKK-RMLSKVLHLYQDLFPHLYDFSPPCWSFPEDRARIEEI--L 64
+RS Q N PG + + +K M + + + + F + P+D + ++
Sbjct 503 IRSHQKYNHIPGSFRIGRKDTMWRSIYNNMKKFGKKEFGFMQKSYIMPDDLESLRQVWPK 562
Query 65 QSSKSETYIIKPDGGAMGSGVQLVSR 90
+SK +I+KP A G+G+++V++
Sbjct 563 NASKLTKWIVKPPASARGTGIRIVNK 588
> 7302451
Length=892
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 47/93 (50%), Gaps = 1/93 (1%)
Query 2 YDHLLFVRSAQIVNRFPGMWDMTKKRMLSKVLHLYQDLFPHLYDFSPPCWSFPEDRARIE 61
+D L ++ Q +N FPGM ++ +K +LS+ L+ +FP Y P W P D +
Sbjct 197 HDLLRNMKRFQQINHFPGMVEICRKDLLSRNLNRMLKMFPGDYRIFPKTWLMPTDAYDV- 255
Query 62 EILQSSKSETYIIKPDGGAMGSGVQLVSRLKDI 94
I + T+I+KP G G+ + + L+ +
Sbjct 256 AIYANKHKRTFILKPYSAGQGRGIWITTDLRTV 288
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 46/96 (47%), Gaps = 6/96 (6%)
Query 98 SNSKRLLSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVYQGEQQQQQQ 157
+ SKR LS K L DV + W S+ + KT + P L+ Y + + Q
Sbjct 377 AGSKRKLSAFNKWLVDHNYDVGEFWASVDDAIIKTLISAWPTLKHNYNVCFPKHDKIQA- 435
Query 158 QQRSRCFQFLGLDILLDADDKCWLLEVNSSPSLRVD 193
FQ LG DIL+D K ++LEVN +PSL D
Sbjct 436 -----SFQLLGFDILVDWKLKPYILEVNHTPSLSAD 466
> Hs11068135
Length=423
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 48/98 (48%), Gaps = 15/98 (15%)
Query 12 QIVNRFPGMWDMTKKRMLSKVLHLYQDLF--------------PHLY-DFSPPCWSFPED 56
QIVN FP +++T+K ++ K + Y+ +LY DF P + P D
Sbjct 60 QIVNHFPNHYELTRKDLMVKNIKRYRKELEKEGSPLAEKDENGKYLYLDFVPVTYMLPAD 119
Query 57 RARIEEILQSSKSETYIIKPDGGAMGSGVQLVSRLKDI 94
E + S S T+I+KP G A G G+ L+++L I
Sbjct 120 YNLFVEEFRKSPSSTWIMKPCGKAQGKGIFLINKLSQI 157
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 163 CFQFLGLDILLDADDKCWLLEVNSSPSL 190
CF+ G DI++D K WL+EVN+SPSL
Sbjct 306 CFECYGYDIIIDDKLKPWLIEVNASPSL 333
> Hs7661970
Length=1199
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 43/86 (50%), Gaps = 17/86 (19%)
Query 104 LSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPW----LELRYRHVYQGEQQQQQQQQ 159
L ++ L +GV+ D +W+ IK++ KT + +P+ L++ R Y
Sbjct 836 LKALWNYLSQKGVNSDAIWEKIKDVVVKTIISSEPYVTSLLKMYVRRPYS---------- 885
Query 160 RSRCFQFLGLDILLDADDKCWLLEVN 185
C + G DI+LD + K W+LEVN
Sbjct 886 ---CHELFGFDIMLDENLKPWVLEVN 908
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 43/84 (51%), Gaps = 1/84 (1%)
Query 8 VRSAQIVNRFPGMWDMTKKRMLSKVLHLYQDLFPHL-YDFSPPCWSFPEDRARIEEILQS 66
+R Q +N FPG + + +K L + L Q F + F P + P+D + + +S
Sbjct 653 IREHQKLNHFPGSFQIGRKDRLWRNLSRMQSRFGKKEFSFFPQSFILPQDAKLLRKAWES 712
Query 67 SKSETYIIKPDGGAMGSGVQLVSR 90
S + +I+KP A G G+Q++ +
Sbjct 713 SSRQKWIVKPPASARGIGIQVIHK 736
> 7297795
Length=827
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 50/102 (49%), Gaps = 9/102 (8%)
Query 89 SRLKDIHNTSNSKRLLSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVY 148
S+ +D++ K + ++ L GV D +W++++ + +T + + + R
Sbjct 540 SKNEDVNACHGHKWTIKSLWTYLANRGVRTDCLWEALRSLVLRTILAGENGINSMIR--- 596
Query 149 QGEQQQQQQQQRSRCFQFLGLDILLDADDKCWLLEVNSSPSL 190
+ + CF+ G D++LD+D WLLEVN SPSL
Sbjct 597 ------ANVESKYSCFELFGFDVILDSDLVPWLLEVNISPSL 632
Score = 33.5 bits (75), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 43/88 (48%), Gaps = 7/88 (7%)
Query 8 VRSAQIVNRFPGMWDMTKKRMLSKVLHLYQDLFPHL---YDFSPPCWSFPEDRARIEE-- 62
+RS Q +N PG + + +K K +L + + H + F P + P D +
Sbjct 370 IRSYQKINHLPGSFRIGRKDSCWK--NLQRQMGKHSNKEFGFMPRTYIIPNDLGALRRHW 427
Query 63 ILQSSKSETYIIKPDGGAMGSGVQLVSR 90
+ ++ +IIKP A G+G+++++R
Sbjct 428 PKYAQRNTKWIIKPPASARGAGIRVINR 455
> 7301272
Length=917
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 44/91 (48%), Gaps = 11/91 (12%)
Query 104 LSDVFKDLKAEGVDVDKVWKSIKEITAKT-FVCLQPWLELRYRHVYQGEQQQQQQQQRSR 162
LS + + LK + D ++ +I+++ K C Q + V G +
Sbjct 447 LSALLRHLKLQSCDTRQLMLNIEDLIIKAVLACAQSIISACRMFVPNG----------NN 496
Query 163 CFQFLGLDILLDADDKCWLLEVNSSPSLRVD 193
CF+ G DIL+D K WLLE+N SPS+ VD
Sbjct 497 CFELYGFDILIDNALKPWLLEINLSPSMGVD 527
Score = 32.3 bits (72), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 39/81 (48%), Gaps = 6/81 (7%)
Query 12 QIVNRFPGMWDMTKKRMLSKVLHLYQDL--FPHLYDFSPPCWSFP-EDRARIEEILQSSK 68
Q VN FP ++MT+K L K + Q L H +D P + P E R + + +
Sbjct 268 QRVNHFPRSYEMTRKDRLYKNIERMQHLRGMKH-FDIVPQTFVLPIESRDLV--VAHNKH 324
Query 69 SETYIIKPDGGAMGSGVQLVS 89
+I+KP + G G+ +V+
Sbjct 325 RGPWIVKPAASSRGRGIFIVN 345
> Hs13994343
Length=592
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 42/90 (46%), Gaps = 17/90 (18%)
Query 104 LSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVYQGEQQQQQQQQRSRC 163
LS F L++ VD +WK I + T + + P + + C
Sbjct 324 LSRFFSYLRSWDVDDLLLWKKIHRMVILTILAIAPSVPFA-----------------ANC 366
Query 164 FQFLGLDILLDADDKCWLLEVNSSPSLRVD 193
F+ G DIL++ + K WLLEVN SP+L +D
Sbjct 367 FELFGFDILINDNLKPWLLEVNYSPALTLD 396
Score = 36.2 bits (82), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 42/94 (44%), Gaps = 7/94 (7%)
Query 8 VRSAQIVNRFPGMWDMTKKRMLSKVL-HLYQDLFPHLYDFSPPCWSFPEDRARI------ 60
V+ Q +N PG +T+K L+K L H+ + LY F P + P D +
Sbjct 138 VKPWQQLNHHPGTTKLTRKDCLAKHLKHMRRMYGTSLYQFIPLTFVMPNDYTKFVAEYFQ 197
Query 61 EEILQSSKSETYIIKPDGGAMGSGVQLVSRLKDI 94
E + +K +I KP + G G+ + S KD
Sbjct 198 ERQMLGTKHSYWICKPAELSRGRGILIFSDFKDF 231
> Hs18572403
Length=245
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 33/64 (51%), Gaps = 6/64 (9%)
Query 37 QDLFPHLYDFSPPCWSFPEDRARIEEILQSSKSE------TYIIKPDGGAMGSGVQLVSR 90
Q+LFP Y+F P W P++ +Q K + T+I+KPDGG G G+ L+
Sbjct 2 QNLFPEEYNFYPRSWILPDEFQLFVAQVQMVKDDDPSWKPTFIVKPDGGCQGDGIYLIKD 61
Query 91 LKDI 94
DI
Sbjct 62 PSDI 65
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 29/49 (59%), Gaps = 0/49 (0%)
Query 97 TSNSKRLLSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYR 145
++ SKR S + L ++GVD+ KVW I + KT + L P L++ Y+
Sbjct 158 STGSKRTFSSILCRLSSKGVDIKKVWSDIISVVIKTVIALTPELKVFYQ 206
> 7293324
Length=496
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/116 (25%), Positives = 52/116 (44%), Gaps = 25/116 (21%)
Query 3 DHLLFVRSAQIVNRFPGMWDMTKKRMLSKVLHLY-QDL---------------------- 39
DH +RS Q++N FP ++++K +L K + Y +DL
Sbjct 103 DHPYRMRSDQVINHFPNSIELSRKDLLIKNIKRYRKDLERRGDPLAQSHPPDTKLGIGGT 162
Query 40 -FPHLYDFSPPCWSFPEDRARIEEILQSSKSETYIIKPDGGAMGSGVQLVSRLKDI 94
+ HL D P + P D E+ + + T+I+KP + G G+ LV++L +
Sbjct 163 RYKHL-DIIPMTFVLPSDYQMFVEVFHRNPASTWIVKPCSKSQGVGIYLVNKLSKL 217
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 18/80 (22%)
Query 111 LKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVYQGEQQQQQQQQRSRCFQFLGLD 170
L+ EGV D +W I + + P + RH CF+ G D
Sbjct 328 LRGEGVS-DMLWSRITATIRHSLDAVAPVMA-NDRH----------------CFEVYGYD 369
Query 171 ILLDADDKCWLLEVNSSPSL 190
I++D + K WL+E+N+SPS+
Sbjct 370 IIIDNNLKPWLIEINTSPSM 389
> Hs13375958
Length=572
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 19/33 (57%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 161 SRCFQFLGLDILLDADDKCWLLEVNSSPSLRVD 193
S CF+ LG DILLD K WLLE+N +PS D
Sbjct 12 SVCFEVLGFDILLDRKLKPWLLEINRAPSFGTD 44
> 7292526
Length=487
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 49/103 (47%), Gaps = 20/103 (19%)
Query 12 QIVNRFPGMWDMTKKRMLSKVLHLYQ-----DLFP--------------HLY-DFSPPCW 51
Q++N FP +++++K +L K + Y+ D P +LY DF P +
Sbjct 131 QMINHFPNHYELSRKDLLVKNIKRYRKDLERDGNPLAEKTESNNSSGTRYLYLDFVPTTF 190
Query 52 SFPEDRARIEEILQSSKSETYIIKPDGGAMGSGVQLVSRLKDI 94
P D E + T+I+KP G + G+G+ L+++L +
Sbjct 191 VLPADYNMFVEEYRKFPLSTWIMKPCGKSQGAGIFLINKLSKL 233
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 163 CFQFLGLDILLDADDKCWLLEVNSSPSL 190
CF+ G DI++D K WL+EVN+SPSL
Sbjct 381 CFECYGYDIIIDNALKPWLVEVNASPSL 408
> CE23614_2
Length=614
Score = 38.1 bits (87), Expect = 0.011, Method: Composition-based stats.
Identities = 28/95 (29%), Positives = 43/95 (45%), Gaps = 13/95 (13%)
Query 101 KRLLSDVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWLELRYRHVYQGEQQQQQQQQR 160
K L + + ++ EG D + I+++ K+ + +Q + R R
Sbjct 248 KWTLGALLRYVENEGKDAKLLMLRIEDLIVKSLLSIQNSVATASR-----------TNLR 296
Query 161 SRC--FQFLGLDILLDADDKCWLLEVNSSPSLRVD 193
C F+ G D+L+D K WLLEVN SPSL D
Sbjct 297 FACTNFELFGFDVLVDQALKPWLLEVNLSPSLACD 331
Score = 36.2 bits (82), Expect = 0.039, Method: Composition-based stats.
Identities = 26/111 (23%), Positives = 46/111 (41%), Gaps = 14/111 (12%)
Query 12 QIVNRFPGMWDMTKKRMLSKVLHLYQDLFPHLYDFSPPCWSFPEDRARIEEILQSSKSET 71
Q +N+FP ++TKK L + + + +F +DF P + P + ++E E
Sbjct 59 QRLNQFPRSTELTKKDRLYENIERSKSIFGESFDFIPEFYVTPRENRKMENAFVRVAKEI 118
Query 72 ------------YIIKPDGGAMGSGVQLVSRLKDIHNTSNSKRLLSDVFKD 110
+I+KP G G+ + + DI + L+S KD
Sbjct 119 AAAGGELCFPGEFIVKPTNSRQGKGIFFANSMADI--PAEGPLLVSRYLKD 167
> SPAC12B10.04
Length=403
Score = 36.2 bits (82), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 158 QQRSRCFQFLGLDILLDADDKCWLLEVNSSPSLR 191
Q CF+ G+D L+D + + +LLEVNS P +
Sbjct 320 QPLENCFEIFGVDFLVDCESQVYLLEVNSYPDFK 353
> Hs20532667_3
Length=145
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 164 FQFLGLDILLDADDKCWLLEVNSSPS 189
FQ G D ++D + K WL+EVN +P+
Sbjct 117 FQLFGFDFMVDEELKVWLIEVNGAPA 142
> Hs20560164
Length=222
Score = 34.7 bits (78), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 30/123 (24%), Positives = 51/123 (41%), Gaps = 9/123 (7%)
Query 1 DYDHLLFVRSAQIVNRFPGMWDMTKKRMLSKVLHLYQD--------LFPHLYDFSPPCWS 52
++DH +I + F +++T+K + K L ++ L DF P +
Sbjct 36 NFDHTYMDEHVRI-SHFRNHYELTRKNYMVKNLKRFRKQLEREAGKLEAAKCDFFPKTFE 94
Query 53 FPEDRARIEEILQSSKSETYIIKPDGGAMGSGVQLVSRLKDIHNTSNSKRLLSDVFKDLK 112
P + E + + T+I+KP + G G+ L RLKDI + R D D+
Sbjct 95 MPCEYHLFVEEFRKNPGITWIMKPVARSQGKGIFLFRRLKDIVDWRKDTRSSDDQKDDIP 154
Query 113 AEG 115
E
Sbjct 155 VEN 157
> 7297165
Length=992
Score = 33.1 bits (74), Expect = 0.42, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 154 QQQQQQRSRCFQFLGLDILLDADDKCWLLEVNSSPSL 190
Q+ +R F+ G D ++ + WL+E+NSSP L
Sbjct 575 QENMDRRPNTFELFGADFMICENFYPWLIEINSSPDL 611
> Hs7661564
Length=352
Score = 32.3 bits (72), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 147 VYQGEQQQQQQQQRSRCFQFLGLDILLDADDKCWLLEVNSSPSL 190
++ + Q Q R F+ G D + D + WL+E+N+SP++
Sbjct 221 IHALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASPTM 264
> CE24779
Length=1781
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 30/129 (23%), Positives = 56/129 (43%), Gaps = 14/129 (10%)
Query 6 LFVRSAQIVNRFPGMW--DMTKKRMLSKVL-----HLYQDLFPHLYDFSPPCWSFPEDRA 58
L +++ NR P DM K ++S L + PHL+ P DR
Sbjct 348 LSLKAYHATNRIPKNLSNDMETKLLISTQLVPGNVSQIKARHPHLFSRCPAAILRRADRT 407
Query 59 RIEEILQSSKSETYI--IKPDGGAMGSGVQLVSRLKDIHNTSNSKRLLSDVFKDLKAEGV 116
+ ++ S++E YI ++ + S + +RL H ++ ++ +VF+ + G
Sbjct 408 AVS--IEDSRNEMYITLMQAELSGKSSDRNIEARL---HVVESNGHVMENVFETISVTGS 462
Query 117 DVDKVWKSI 125
+ V+KSI
Sbjct 463 QLSTVYKSI 471
> HsM19923582
Length=1749
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 23/37 (62%), Gaps = 4/37 (10%)
Query 158 QQRSRCFQFLGLDILLDA----DDKCWLLEVNSSPSL 190
Q+R R + +G+ + + DDKC+L+ +N+ P+L
Sbjct 422 QERQRQLESMGISLEMSGIKVGDDKCYLVNLNADPAL 458
> Hs21361722
Length=1805
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 23/37 (62%), Gaps = 4/37 (10%)
Query 158 QQRSRCFQFLGLDILLDA----DDKCWLLEVNSSPSL 190
Q+R R + +G+ + + DDKC+L+ +N+ P+L
Sbjct 422 QERQRQLESMGISLEMSGIKVGDDKCYLVNLNADPAL 458
> 7299369
Length=516
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 164 FQFLGLDILLDADDKCWLLEVNSSPSLRVDYF 195
F + D+ +D D K +L+E N SP+L +F
Sbjct 346 FDLMRFDLFIDEDLKVFLMEANMSPNLSSAHF 377
> Hs19115954
Length=4624
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 48/109 (44%), Gaps = 15/109 (13%)
Query 85 VQLVSRLKDIHNTSNSKRLLS-DVFKDLKAEGVDVDKVWKSIKEITAKTFVCLQPWL-EL 142
V+LVS L I N++ + + S D FK + +W+ KE KTF+ P L E
Sbjct 1117 VKLVSVLSTIINSTKKEVITSMDCFKR-------YNHIWQKGKEEAIKTFITQSPLLSEF 1169
Query 143 RYRHVY-QGEQQQQQQQQRSRCFQFLG-----LDILLDADDKCWLLEVN 185
+ +Y Q +Q+ + C + L L A+ K W++ +
Sbjct 1170 ESQILYFQNLEQEINAEPEYVCVGSIALYTADLKFALTAETKAWMVVIG 1218
> Hs6005922_1
Length=2559
Score = 29.3 bits (64), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 11/19 (57%), Positives = 13/19 (68%), Gaps = 0/19 (0%)
Query 41 PHLYDFSPPCWSFPEDRAR 59
P L D +PPCWS + RAR
Sbjct 2283 PSLSDTTPPCWSPLQPRAR 2301
> Hs18549045
Length=235
Score = 29.3 bits (64), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 75 KPDGGAMGSGVQLVSRLKDIHNTSNSKRLLS 105
K G A G G+ ++ +LKD+ ++ NS+ LLS
Sbjct 19 KTRGQADGFGLDILPKLKDVKSSDNSRSLLS 49
> Hs10835121
Length=574
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 38/69 (55%), Gaps = 5/69 (7%)
Query 9 RSAQIVNRF---PGMWDMTKKRMLSKVLHLYQDLFPHLYDFSP-PCWSFPEDRARIEEIL 64
RSAQ+++R+ + +T+ ++ +HL + +FP LY P W+ DR R++ +
Sbjct 479 RSAQLLSRYRPRAAVIAVTRSAQAARQVHLCRGVFPLLYREPPEAIWADDVDR-RVQFGI 537
Query 65 QSSKSETYI 73
+S K ++
Sbjct 538 ESGKLRGFL 546
> Hs5453774
Length=373
Score = 28.9 bits (63), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 6/63 (9%)
Query 19 GMWDMTKKRMLSKVLHLYQDLFPHLYDFSPPCWSFPEDRARIEEILQSSKSETYIIKPDG 78
G D T + M ++ LY+D+F HL D S +S PE+ A LQ + T + P G
Sbjct 313 GEADRTLEVMRQQLTELYRDIFQHLRDESGNSYS-PEEYA-----LQQAADGTIFLVPRG 366
Query 79 GAM 81
M
Sbjct 367 TKM 369
> CE01399
Length=479
Score = 28.5 bits (62), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 41/87 (47%), Gaps = 12/87 (13%)
Query 56 DRARIEEILQSSKSETYIIKPDGGAMGSGVQLVSRLKDIHNTSNSKRLLSDVFKDLKAEG 115
D EEI + +S I+ +++ ++DI+ +SNS L D++K +K+ G
Sbjct 301 DVTTTEEIGKKVRSALQSIRDISSTNEDLNEILKTVRDIYRSSNSATLFQDLYKIIKSNG 360
Query 116 VDVDKVWKSIKEITAKTFVCLQPWLEL 142
S +E+ F+ LQ +E+
Sbjct 361 --------SYREV----FLALQMLMEM 375
> CE11662
Length=1193
Score = 28.5 bits (62), Expect = 9.1, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 31/51 (60%), Gaps = 3/51 (5%)
Query 81 MGSGVQLVSRLKDIHNTSNSKRLLSDVFKDLKAEGVDVDKVWKSIKEITAK 131
+G G+QL+ +L ++ +N K++++DV K + +V K+ K++ T K
Sbjct 615 LGKGLQLLRQLSEV---NNKKKIVTDVTKFSDSVFAEVQKILKTVDTSTLK 662
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3443493696
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40