bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
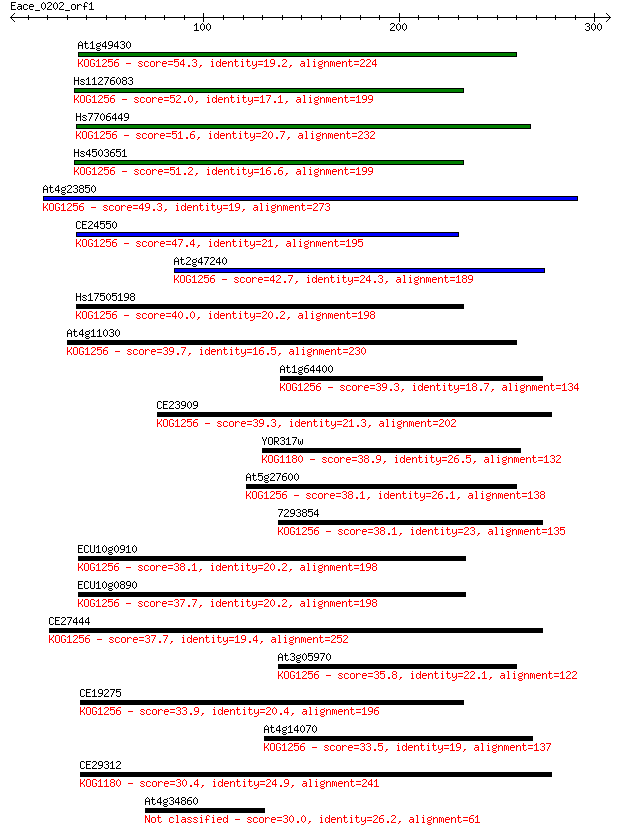
Query= Eace_0202_orf1
Length=307
Score E
Sequences producing significant alignments: (Bits) Value
At1g49430 54.3 3e-07
Hs11276083 52.0 2e-06
Hs7706449 51.6 2e-06
Hs4503651 51.2 3e-06
At4g23850 49.3 1e-05
CE24550 47.4 4e-05
At2g47240 42.7 0.001
Hs17505198 40.0 0.006
At4g11030 39.7 0.008
At1g64400 39.3 0.011
CE23909 39.3 0.011
YOR317w 38.9 0.014
At5g27600 38.1 0.022
7293854 38.1 0.022
ECU10g0910 38.1 0.024
ECU10g0890 37.7 0.032
CE27444 37.7 0.032
At3g05970 35.8 0.13
CE19275 33.9 0.41
At4g14070 33.5 0.52
CE29312 30.4 4.3
At4g34860 30.0 5.8
> At1g49430
Length=666
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 43/227 (18%), Positives = 100/227 (44%), Gaps = 12/227 (5%)
Query 36 GMTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVLDLREGGQLSNLSALITLDAVG 95
G+T VP++ + ++ H +++ + + + +L ++G SNL +++ V
Sbjct 128 GITYVPLYDSLGVNAVEFIINHAEVSLVFVQEKTVSSILSCQKGCS-SNLKTIVSFGEVS 186
Query 96 DQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMPFNG 155
+ A ++L + E + N + E L ++ +I ++Y + G
Sbjct 187 STQKEEAKNQCVSLFSWNEFSLMGN-LDEANLPRKRKT-----DICTIMYTSGTTGEPKG 240
Query 156 AMLTNENV---LASLDTLTDWRARALSIVEADRMLCIDSLASIHQRVLHLAVVDAGATVA 212
+L N + + S+D + + R+ +D LA + +V+ + + G++V
Sbjct 241 VILNNAAISVQVLSIDKMLEVTDRSCDT--SDVFFSYLPLAHCYDQVMEIYFLSRGSSVG 298
Query 213 FARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKSLGRLRR 259
+ R + L DD++A PT+ G + L+A + + + G +R+
Sbjct 299 YWRGDIRYLMDDVQALKPTVFCGVPRVYDKLYAGIMQKISASGLIRK 345
> Hs11276083
Length=698
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/200 (17%), Positives = 86/200 (43%), Gaps = 7/200 (3%)
Query 34 AFGMTVVPMHPEMNARRFLEVMIHTQMTHLCTDW-RHLEIVLDLREGGQLSNLSALITLD 92
A+ M +VP++ + ++ +++ + D +++L+ E + L ++ +D
Sbjct 169 AYSMVIVPLYDTLGNEAITYIVNKAELSLVFVDKPEKAKLLLEGVENKLIPGLKIIVVMD 228
Query 93 AVGDQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMP 152
A G + ++R + + + D +G + +++A + + +
Sbjct 229 AYGSELVERGQRCGVEVTSMKAMED----LGRANRRKPKPPA--PEDLAVICFTSGTTGN 282
Query 153 FNGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLASIHQRVLHLAVVDAGATVA 212
GAM+T+ N+++ ++ D ++ LA + +RV+ ++ GA +
Sbjct 283 PKGAMVTHRNIVSDCSAFVKATENTVNPCPDDTLISFLPLAHMFERVVECVMLCHGAKIG 342
Query 213 FARSGLLTLGDDLEAFHPTI 232
F + + L DDL+ PT+
Sbjct 343 FFQGDIRLLMDDLKVLQPTV 362
> Hs7706449
Length=683
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 48/233 (20%), Positives = 97/233 (41%), Gaps = 11/233 (4%)
Query 35 FGMTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVLDLR-EGGQLSNLSALITLDA 93
+ M VP++ + + ++ + + D +VL E G +L +I +D
Sbjct 155 YSMVAVPLYDTLGPEAIVHIVNKADIAMVICDTPQKALVLIGNVEKGFTPSLKVIILMDP 214
Query 94 VGDQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMPF 153
D QR + ++ L D+ N E K + S ++++ + + +
Sbjct 215 FDDDLKQRGEK---SGIEILSLYDAENLGKEHFRKPVPPS---PEDLSVICFTSGTTGDP 268
Query 154 NGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLASIHQRVLHLAVVDAGATVAF 213
GAM+T++N++++ A D + LA + +R++ V GA V F
Sbjct 269 KGAMITHQNIVSNAAAFLKCVEHAYEPTPDDVAISYLPLAHMFERIVQAVVYSCGARVGF 328
Query 214 ARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKSLGRLRRFWVLLKL 266
+ + L DD++ PT+ +L ++ + K+ L++F LLKL
Sbjct 329 FQGDIRLLADDMKTLKPTLFPAVPRLLNRIYDKVQNEAKT--PLKKF--LLKL 377
> Hs4503651
Length=699
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/200 (16%), Positives = 86/200 (43%), Gaps = 7/200 (3%)
Query 34 AFGMTVVPMHPEMNARRFLEVMIHTQMTHLCTDW-RHLEIVLDLREGGQLSNLSALITLD 92
A+ M +VP++ + ++ +++ + D +++L+ E + L ++ +D
Sbjct 169 AYSMVIVPLYDTLGNEAITYIVNKAELSLVFVDKPEKAKLLLEGVENKLIPGLKIIVVMD 228
Query 93 AVGDQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMP 152
+ G + ++R + + + D +G + +++A + + +
Sbjct 229 SYGSELVERGQRCGVEVTSMKAMED----LGRANRRKPKPPA--PEDLAVICFTSGTTGN 282
Query 153 FNGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLASIHQRVLHLAVVDAGATVA 212
GAM+T+ N+++ ++ D ++ LA + +RV+ ++ GA +
Sbjct 283 PKGAMVTHRNIVSDCSAFVKATENTVNPCPDDTLISFLPLAHMFERVVECVMLCHGAKIG 342
Query 213 FARSGLLTLGDDLEAFHPTI 232
F + + L DDL+ PT+
Sbjct 343 FFQGDIRLLMDDLKVLQPTV 362
> At4g23850
Length=666
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 52/279 (18%), Positives = 116/279 (41%), Gaps = 19/279 (6%)
Query 18 WHAHAHKHAQARKTAGAFGMTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVLDLR 77
+ A++ + + + A G+ VP++ + A ++ H++++ + + + + +
Sbjct 110 YGANSPEWIISMEACNAHGLYCVPLYDTLGADAVEFIISHSEVSIVFVEEKKISELFKTC 169
Query 78 EGGQLSNLSALITLDAVGDQAMQRAHALDLTLV---DFWELADSPNGIGEQRLKSLHRSG 134
+ +++ V + + A L + +F +L G G+Q + +
Sbjct 170 PN-STEYMKTVVSFGGVSREQKEEAETFGLVIYAWDEFLKL-----GEGKQYDLPIKKK- 222
Query 135 FDTQEIAAVIYPRALSMPFNGAMLTNENV---LASLDTLTDWRARALSIVEADRMLCIDS 191
+I ++Y + G M++NE++ +A + L AL++ D L
Sbjct 223 ---SDICTIMYTSGTTGDPKGVMISNESIVTLIAGVIRLLKSANEALTV--KDVYLSYLP 277
Query 192 LASIHQRVLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTL 251
LA I RV+ + GA + F R + L +DL PTI +L +++ ++ L
Sbjct 278 LAHIFDRVIEECFIQHGAAIGFWRGDVKLLIEDLAELKPTIFCAVPRVLDRVYSGLQKKL 337
Query 252 KSLGRLRRFWVLLKLRWRARQYWQTGDFELLGKPPFEEL 290
G L++F ++ Y + G + P F++L
Sbjct 338 SDGGFLKKFIFDSAFSYKF-GYMKKGQSHVEASPLFDKL 375
> CE24550
Length=719
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 41/196 (20%), Positives = 84/196 (42%), Gaps = 9/196 (4%)
Query 35 FGMTVVPMHPEMNARRFLEVMIHTQMT-HLCTDWRHLEIVLDLREGGQLSNLSALITLDA 93
+ +VP++ + + + ++ ++ +C D +L +E Q +LS L+ ++
Sbjct 193 YSNVIVPIYETLGSEASIFILNQAEIKIVVCDDISKATGLLKFKE--QCPSLSTLVVMEP 250
Query 94 VGDQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMPF 153
V D+ A +L + ++ F +L IG+ +++A + Y +
Sbjct 251 VTDELKTTASSLGVEVLTFEDLEK----IGKNAKTRPAHIPPTPEDLATICYTSGTTGTP 306
Query 154 NGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLASIHQRVLHLAVVDAGATVAF 213
G MLT+ NV+A + ++ I D M+ LA + +RV+ GA V F
Sbjct 307 KGVMLTHANVIADGVCMDFFKHSG--IAATDSMISFLPLAHMLERVIESVCFCVGAKVGF 364
Query 214 ARSGLLTLGDDLEAFH 229
R + L +D++
Sbjct 365 YRGDIRVLAEDIKELK 380
> At2g47240
Length=660
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 46/199 (23%), Positives = 77/199 (38%), Gaps = 21/199 (10%)
Query 85 LSALITLDAVGDQAMQRAHALDL---TLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIA 141
L A+++ V D+ +A + + + +DF + R K + I
Sbjct 173 LKAIVSFTNVSDELSHKASEIGVKTYSWIDFLHMG---------REKPEDTNPPKAFNIC 223
Query 142 AVIYPRALSMPFNGAMLTNENV---LASLDTLTDWRARALSIVEADRMLCIDSLASIHQR 198
++Y S G +LT++ V + +D D ++ D L LA I R
Sbjct 224 TIMYTSGTSGDPKGVVLTHQAVATFVVGMDLYMDQFEDKMT--HDDVYLSFLPLAHILDR 281
Query 199 VLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKSLGRLR 258
+ GA+V + L L DD++ PT +AG + +H ++ L+ L R
Sbjct 282 MNEEYFFRKGASVGYYHGNLNVLRDDIQELKPTYLAGVPRVFERIHEGIQKALQELNPRR 341
Query 259 RFWVLL----KLRWRARQY 273
RF KL W R Y
Sbjct 342 RFIFNALYKHKLAWLNRGY 360
> Hs17505198
Length=697
Score = 40.0 bits (92), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 40/201 (19%), Positives = 81/201 (40%), Gaps = 11/201 (5%)
Query 35 FGMTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVL-DLREGGQLSNLSALITLDA 93
+ M VVP++ + ++ ++ + D ++L + E + L +I +D
Sbjct 170 YSMVVVPLYDTLGPGAIRYIINTADISTVIVDKPQKAVLLLEHVERKETPGLKLIILMDP 229
Query 94 VGDQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQ--EIAAVIYPRALSM 151
+ +R + + + D G++ H++ Q +++ V + +
Sbjct 230 FEEALKERGQKCGVVIKSMQAVEDC----GQEN----HQAPVPPQPDDLSIVCFTSGTTG 281
Query 152 PFNGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLASIHQRVLHLAVVDAGATV 211
GAMLT+ NV+A + AD + LA + +R++ V G V
Sbjct 282 NPKGAMLTHGNVVADFSGFLKVTESQWAPTCADVHISYLPLAHMFERMVQSVVYCHGGRV 341
Query 212 AFARSGLLTLGDDLEAFHPTI 232
F + + L DD++A PTI
Sbjct 342 GFFQGDIRLLSDDMKALCPTI 362
> At4g11030
Length=666
Score = 39.7 bits (91), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 38/234 (16%), Positives = 93/234 (39%), Gaps = 14/234 (5%)
Query 30 KTAGAFGMTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVLDLREGGQLSNLSALI 89
+ A G+ VP++ + A ++ H +++ + + + + + ++
Sbjct 122 EACNAHGLYCVPLYDTLGAGAVEFIISHAEVSIAFVEEKKIPELFKTCPN-STKYMKTVV 180
Query 90 TLDAVGDQAMQRAHALDLTL---VDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYP 146
+ V + + A L L + +F +L G G+Q + + +I ++Y
Sbjct 181 SFGGVKPEQKEEAEKLGLVIHSWDEFLKL-----GEGKQYELPIKKP----SDICTIMYT 231
Query 147 RALSMPFNGAMLTNENVLASLDTLTDWRARA-LSIVEADRMLCIDSLASIHQRVLHLAVV 205
+ G M++NE+++ + + S+ E D + LA + R + ++
Sbjct 232 SGTTGDPKGVMISNESIVTITTGVMHFLGNVNASLSEKDVYISYLPLAHVFDRAIEECII 291
Query 206 DAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKSLGRLRR 259
G ++ F R + L +DL P+I +L ++ ++ L G ++
Sbjct 292 QVGGSIGFWRGDVKLLIEDLGELKPSIFCAVPRVLDRVYTGLQQKLSGGGFFKK 345
> At1g64400
Length=665
Score = 39.3 bits (90), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 25/135 (18%), Positives = 58/135 (42%), Gaps = 1/135 (0%)
Query 139 EIAAVIYPRALSMPFNGAMLTNENVLASLDTLTDW-RARALSIVEADRMLCIDSLASIHQ 197
++ ++Y + G +LTNE+++ L+ + + + D L LA I
Sbjct 224 DVCTIMYTSGTTGDPKGVLLTNESIIHLLEGVKKLLKTIDEELTSKDVYLSYLPLAHIFD 283
Query 198 RVLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKSLGRL 257
RV+ + A++ F R + L +D+ A PT+ +L ++ ++ L G +
Sbjct 284 RVIEELCIYEAASIGFWRGDVKILIEDIAALKPTVFCAVPRVLERIYTGLQQKLSDGGFV 343
Query 258 RRFWVLLKLRWRARQ 272
++ +++ +
Sbjct 344 KKKLFNFAFKYKHKN 358
> CE23909
Length=700
Score = 39.3 bits (90), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 43/204 (21%), Positives = 83/204 (40%), Gaps = 13/204 (6%)
Query 76 LREGGQLSNLSALITLDAVGDQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGF 135
+R+ LS+L ++ + D + A D L F E E K HR
Sbjct 204 IRDKSYLSSLKYIVQFNECSDDIKEMARENDFRLWSFNEFV-------EMGKKQKHRPHV 256
Query 136 --DTQEIAAVIYPRALSMPFNGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLA 193
+ +A + + + G MLT+ N+ ++ + ++ A D L LA
Sbjct 257 PPTPETLATISFTSGTTGRPKGVMLTHLNMCSATMSCEEFENEAGV---QDAYLSYLPLA 313
Query 194 SIHQRVLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKS 253
I++R+ L+ G+ + F+R L DD++A P A ++ +H A + ++
Sbjct 314 HIYERLCLLSNFMIGSRIGFSRGDPKLLVDDVQALAPRSFATVPRVIDKIHKAVMKQVQD 373
Query 254 LGRLRRFWVLLKLRWRARQYWQTG 277
L++ + + ++ Y TG
Sbjct 374 -KPLKKMILNAAIAYKLYHYKMTG 396
> YOR317w
Length=700
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 35/141 (24%), Positives = 58/141 (41%), Gaps = 14/141 (9%)
Query 130 LHRSGFDTQEIAAVIYPRALSMPFNGAMLTNENVLASLDTLTDWRARALSIV-EADRMLC 188
+H G D ++ ++Y + G +L + NV+A + + L V DR++C
Sbjct 256 VHPPGKD--DLCCIMYTSGSTGEPKGVVLKHSNVVAGVGGAS---LNVLKFVGNTDRVIC 310
Query 189 IDSLASIHQRVLHLAVVDAGATVAFARSGLLTLGD------DLEAFHPTIVAGSGGILPV 242
LA I + V L GA + +A LT DL+ F PTI+ G +
Sbjct 311 FLPLAHIFELVFELLSFYWGACIGYATVKTLTSSSVRNCQGDLQEFKPTIMVGVAAVWET 370
Query 243 LHAAARRTLKSLGRLRR--FW 261
+ + +L L + FW
Sbjct 371 VRKGILNQIDNLPFLTKKIFW 391
> At5g27600
Length=698
Score = 38.1 bits (87), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 61/149 (40%), Gaps = 20/149 (13%)
Query 122 IGEQRLKSLHRSGF------DTQEIAAVIYPRALSMPFNGAMLTNENVLASLDTLTDWRA 175
+ Q+L S RS ++IA + Y + G +LT+ N++A++
Sbjct 237 VSYQKLLSQGRSSLHPFSPPKPEDIATICYTSGTTGTPKGVVLTHGNLIANV-------- 288
Query 176 RALSIVEA-----DRMLCIDSLASIHQRVLHLAVVDAGATVAFARSGLLTLGDDLEAFHP 230
A S VEA D + LA I++R + V G V F + + L DD P
Sbjct 289 -AGSSVEAEFFPSDVYISYLPLAHIYERANQIMGVYGGVAVGFYQGDVFKLMDDFAVLRP 347
Query 231 TIVAGSGGILPVLHAAARRTLKSLGRLRR 259
TI + ++ +KS G +++
Sbjct 348 TIFCSVPRLYNRIYDGITSAVKSSGVVKK 376
> 7293854
Length=704
Score = 38.1 bits (87), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 31/139 (22%), Positives = 61/139 (43%), Gaps = 10/139 (7%)
Query 138 QEIAAVIYPRALSMPFNGAMLTNENVLASLDT----LTDWRARALSIVEADRMLCIDSLA 193
+++ V Y + G MLT+ NV+A + + + D R RA D M+ LA
Sbjct 275 EDLCTVCYTSGTTGNPKGVMLTHGNVVAGVCSVILQMGDHRIRA-----GDVMVSFLPLA 329
Query 194 SIHQRVLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKS 253
+ +R + G V F + L +DL+ PT++ +L ++ + + +
Sbjct 330 HMFERCCENGMYYVGGCVGFYSGDIKELTNDLKMLKPTVMPAVPRLLNRVYDKIQNDISA 389
Query 254 LGRLRRFWVLLKLRWRARQ 272
G L+R + +R + ++
Sbjct 390 SG-LKRGLFNMAMRAKEKE 407
> ECU10g0910
Length=708
Score = 38.1 bits (87), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 40/201 (19%), Positives = 81/201 (40%), Gaps = 12/201 (5%)
Query 36 GMTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVLDLREGGQLSNLSALITLDAVG 95
G VP++ + +++ T+M +++L+ + LS ++ +D
Sbjct 191 GCINVPLYSTFSPADVRQILAETEMKVCVASADKAQLLLETVLEDSKAGLSCIVLMDRDA 250
Query 96 DQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMPFNG 155
A +R + + F ++ S + G +R ++A + Y S G
Sbjct 251 GVA-ERLRNAGVLVYYFGDVVSSGDKAGRKRYPK-------GDDLATICYTSGTSGSPKG 302
Query 156 AMLTNENVLASLDTLTDWRARALSIVEADR---MLCIDSLASIHQRVLHLAVVDAGATVA 212
MLT+ N ++ + ++ +RA +VE R L LA + +RV + +G +
Sbjct 303 VMLTHRNFVSCVAGISRG-SRAGEMVEIGRDDVYLSYLPLAHVMERVCMNISMASGCRIV 361
Query 213 FARSGLLTLGDDLEAFHPTIV 233
F R L + +D PT +
Sbjct 362 FFRGNLKEIKEDYLVVRPTFI 382
> ECU10g0890
Length=626
Score = 37.7 bits (86), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 40/201 (19%), Positives = 81/201 (40%), Gaps = 12/201 (5%)
Query 36 GMTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVLDLREGGQLSNLSALITLDAVG 95
G VP++ + +++ T+M +++L+ + LS ++ +D
Sbjct 109 GCINVPLYSTFSPADVRQILAETEMKVCVASADKAQLLLETVLEDSKAGLSCIVLMDRDA 168
Query 96 DQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMPFNG 155
A +R + + F ++ S + G +R ++A + Y S G
Sbjct 169 GVA-ERLRNAGVLVYYFGDVVSSGDKAGRKRYPK-------GDDLATICYTSGTSGSPKG 220
Query 156 AMLTNENVLASLDTLTDWRARALSIVEADR---MLCIDSLASIHQRVLHLAVVDAGATVA 212
MLT+ N ++ + ++ +RA +VE R L LA + +RV + +G +
Sbjct 221 VMLTHRNFVSCVAGISRG-SRAGEMVEIGRDDVYLSYLPLAHVMERVCMNISMASGCRIV 279
Query 213 FARSGLLTLGDDLEAFHPTIV 233
F R L + +D PT +
Sbjct 280 FFRGNLKEIKEDYLVVRPTFI 300
> CE27444
Length=653
Score = 37.7 bits (86), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 49/259 (18%), Positives = 99/259 (38%), Gaps = 21/259 (8%)
Query 21 HAHKHAQARKTAGAFGMTVVPMHPEMNARRFLEVMIHTQMTHL-CTDWRHLEIVLDLREG 79
H+ K+ F +T VP++ + +++ + ++ + C + + E L + G
Sbjct 108 HSRKYMHTMHGISGFDLTTVPLYHQSKLETLCDIIDNCKLEIIFCENAKRAEGFLSSKLG 167
Query 80 GQLSNLSALITLDAVGDQAMQRAHALD-LTLVDFWELA--DSPNGIG---EQRLKSLHRS 133
+L +L LI LD +Q+ ++ ++L +F E+ + N + E H S
Sbjct 168 DRLRSLKTLIILDKT--TTLQKHDDVEVMSLDEFKEIGKRNKRNPVKPKPETTYVICHTS 225
Query 134 GFDTQEIAAVIYPRALSMPFNGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLA 193
G + P+ + M + + + S W+ D SLA
Sbjct 226 GTTGR-------PKGVEMSHGSLVTSVSGIFTSWTIAYKWKFGL-----EDTYFSFLSLA 273
Query 194 SIHQRVLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKS 253
I++ ++ + G + TL ++ PTIV+ +L L+ A +
Sbjct 274 HIYEHLIQTLTIYFGGKIGIYDGNTATLIPQIQKLEPTIVSLVPRLLNKLYEAVHDGVAK 333
Query 254 LGRLRRFWVLLKLRWRARQ 272
G + R + +ARQ
Sbjct 334 KGFIARKLFDYAKKTKARQ 352
> At3g05970
Length=657
Score = 35.8 bits (81), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 27/122 (22%), Positives = 55/122 (45%), Gaps = 4/122 (3%)
Query 138 QEIAAVIYPRALSMPFNGAMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLASIHQ 197
++A + Y + G +LT+ N++A++ + ++ +D + LA I++
Sbjct 246 DDVATICYTSGTTGTPKGVVLTHANLIANVAG----SSFSVKFFSSDVYISYLPLAHIYE 301
Query 198 RVLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARRTLKSLGRL 257
R + V G V F + + L DDL A PT+ + + ++A +K+ G L
Sbjct 302 RANQILTVYFGVAVGFYQGDNMKLLDDLAALRPTVFSSVPRLYNRIYAGIINAVKTSGGL 361
Query 258 RR 259
+
Sbjct 362 KE 363
> CE19275
Length=687
Score = 33.9 bits (76), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 40/199 (20%), Positives = 93/199 (46%), Gaps = 11/199 (5%)
Query 37 MTVVPMHPEMNARRFLEVMIHTQMTHLCTD-WRHLEIVLDLREGGQLSNLSALITLDAVG 95
M VVP++ + A ++ +++ + D ++ E ++ RE + L +I +D+
Sbjct 157 MVVVPLYDTLGAEAATFIISQAEISVVIVDSFKKAESLIKNRE--NMPTLKNIIVIDS-A 213
Query 96 DQAMQRAHALDLTLVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMPFNG 155
D+ +D V+ L ++ N +G + + + D I + Y + G
Sbjct 214 DELKDGTAIIDTIRVE--SLTNALN-LGSRYPFTNNLPKPDDNYI--ICYTSGTTGTPKG 268
Query 156 AMLTNENVLASLDT-LTDWRARALSIVEADRM-LCIDSLASIHQRVLHLAVVDAGATVAF 213
MLT+ N++A++ L A S+++A ++ + L+ + +++ H ++ G+ + +
Sbjct 269 VMLTHSNIVANISGFLKILFAFQPSMIDATQVHISYLPLSHMMEQLTHWTLLGFGSKIGY 328
Query 214 ARSGLLTLGDDLEAFHPTI 232
R + L DD++ PT+
Sbjct 329 FRGSIQGLTDDIKTLKPTV 347
> At4g14070
Length=698
Score = 33.5 bits (75), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 26/138 (18%), Positives = 60/138 (43%), Gaps = 7/138 (5%)
Query 131 HRSGF-DTQEIAAVIYPRALSMPFNGAMLTNENVLASLDTLTDWRARALSIVEADRMLCI 189
+R+ F D+ + AA++Y + G MLT+ N+L + L+ + + D+ L +
Sbjct 242 YRNQFIDSDDTAAIMYTSGTTGNPKGVMLTHRNLLHQIKHLSKY----VPAQAGDKFLSM 297
Query 190 DSLASIHQRVLHLAVVDAGATVAFARSGLLTLGDDLEAFHPTIVAGSGGILPVLHAAARR 249
++R + G + + + L DDL+ + P + + L++ ++
Sbjct 298 LPSWHAYERASEYFIFTCGVEQMY--TSIRYLKDDLKRYQPNYIVSVPLVYETLYSGIQK 355
Query 250 TLKSLGRLRRFWVLLKLR 267
+ + R+F L ++
Sbjct 356 QISASSAGRKFLALTLIK 373
> CE29312
Length=731
Score = 30.4 bits (67), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 60/254 (23%), Positives = 101/254 (39%), Gaps = 26/254 (10%)
Query 37 MTVVPMHPEMNARRFLEVMIHTQMTHLCTDWRHLEIVLDLREGGQLSNLSALITLDAVGD 96
+T+V ++ + + T+ T L T L ++ L G + L LI V
Sbjct 192 VTIVTVYATLGEDAIAHAIGETEATILVTSSELLPKIVTL--GKKCPTLKTLIYFAPVD- 248
Query 97 QAMQRAHALDLT-LVDFWELADSPNGIGEQRLKSLHRSGFDTQEIAAVIYPRALSMPFNG 155
Q+A A +L D ++ S +G+ + + + S +IA ++Y + G
Sbjct 249 ---QKAPAPELAPFRDQFKHVLSLSGLLTRNQEQVKESTAVKSDIALIMYTSGTTGQPKG 305
Query 156 AMLTNENVLASLDTLTDWRARALSIVEADRMLCIDSLASIHQRVLHLAVVDAGATVAFAR 215
+L ++NV+A+L D I AD + LA I + L + GA V +
Sbjct 306 VILLHQNVVAALLGQGDGVG---IICNADTYIGYLPLAHILELDAELTCLTKGAKVGY-- 360
Query 216 SGLLTLGD-----------DLEAFHPTIVAGSGGILPVLHAAARRTLKSLGRLRRFWVLL 264
S LTL D D A PT++A I+ + A + + R + L
Sbjct 361 SSPLTLHDRASKIQKGTHGDCHALRPTLMAAVPAIMDRIFKAVSEEVAASPRFMQ--ELF 418
Query 265 KLRW-RARQYWQTG 277
KL + R R + G
Sbjct 419 KLNYERKRSRYLEG 432
> At4g34860
Length=571
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 29/61 (47%), Gaps = 13/61 (21%)
Query 70 LEIVLDLREGGQLSNLSALITLDAVGDQAMQRAHALDLTLVDFWELADSPNGIGEQRLKS 129
+ +D+ + G + N+ +L TLD + D +DF +L + P + RL+S
Sbjct 4 FNLSVDVNQNGNIKNVDSLSTLDDIDD-------------IDFAKLLEKPRPLNIDRLRS 50
Query 130 L 130
L
Sbjct 51 L 51
Lambda K H
0.325 0.135 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7025720074
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40