bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0204_orf1
Length=220
Score E
Sequences producing significant alignments: (Bits) Value
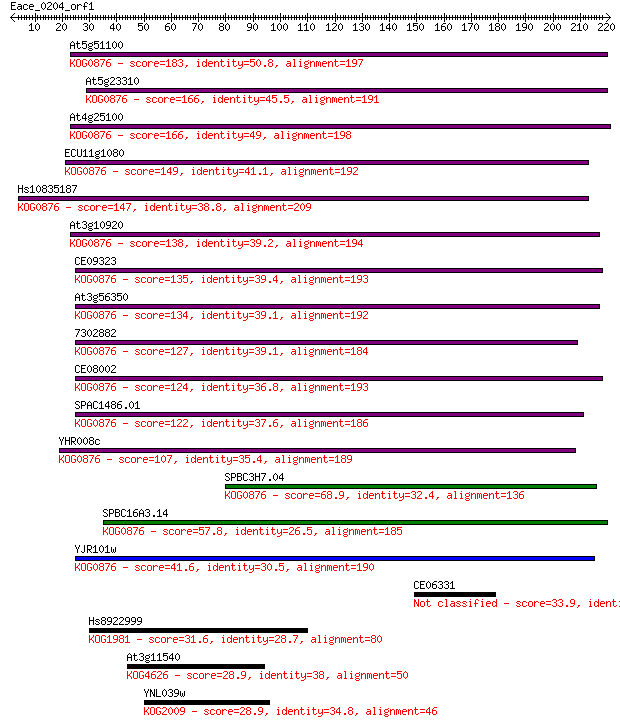
At5g51100 183 2e-46
At5g23310 166 3e-41
At4g25100 166 3e-41
ECU11g1080 149 4e-36
Hs10835187 147 1e-35
At3g10920 138 8e-33
CE09323 135 5e-32
At3g56350 134 1e-31
7302882 127 2e-29
CE08002 124 1e-28
SPAC1486.01 122 6e-28
YHR008c 107 3e-23
SPBC3H7.04 68.9 7e-12
SPBC16A3.14 57.8 2e-08
YJR101w 41.6 0.001
CE06331 33.9 0.25
Hs8922999 31.6 1.4
At3g11540 28.9 7.6
YNL039w 28.9 8.9
> At5g51100
Length=305
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 100/219 (45%), Positives = 132/219 (60%), Gaps = 25/219 (11%)
Query 23 FQLPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASKSLEDLIKTA-- 80
F+L P PY DALEPH+S+ETL+YH+GKHH TYV NLN+ G D + SLE+++ +
Sbjct 53 FELKPPPYPLDALEPHMSRETLDYHWGKHHKTYVENLNKQILGTDLDALSLEEVVLLSYN 112
Query 81 SGGIL---NNAGQVYNHTFYWNSMKAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAG 137
G +L NNA Q +NH F+W S++ GGGG PTG + IE+ F SFE F E+F +AAA
Sbjct 113 KGNMLPAFNNAAQAWNHEFFWESIQPGGGGKPTGELLRLIERDFGSFEEFLERFKSAAAS 172
Query 138 HFGSGWVWLCYCG---------------EQKKVVITQTHDGGVPFR-DNSKCFPLLACDV 181
+FGSGW WL Y E KK+VI +T + P D S PLL D
Sbjct 173 NFGSGWTWLAYKANRLDVANAVNPLPKEEDKKLVIVKTPNAVNPLVWDYS---PLLTIDT 229
Query 182 WEHAYYIDRRNDRAAYVKAWWS-LVNWDFANENLKKAMA 219
WEHAYY+D N RA Y+ + LV+W+ + L+ A+A
Sbjct 230 WEHAYYLDFENRRAEYINTFMEKLVSWETVSTRLESAIA 268
> At5g23310
Length=263
Score = 166 bits (420), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 87/199 (43%), Positives = 127/199 (63%), Gaps = 11/199 (5%)
Query 29 PYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEA--SKSLEDLIKTA--SGGI 84
PY DALEP++S+ TLE H+GKHH YV+NLN+ GKD+ ++E+LIK +G
Sbjct 56 PYPLDALEPYMSRRTLEVHWGKHHRGYVDNLNKQL-GKDDRLYGYTMEELIKATYNNGNP 114
Query 85 L---NNAGQVYNHTFYWNSMKAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAGHFGS 141
L NNA QVYNH F+W SM+ GGG +P + +I+K F SF F+E+F+ AA FGS
Sbjct 115 LPEFNNAAQVYNHDFFWESMQPGGGDTPQKGVLEQIDKDFGSFTNFREKFTNAALTQFGS 174
Query 142 GWVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRNDRAAYVKAW 201
GWVWL E++++ + +T + P + P++ DVWEH+YY+D +NDRA Y+ +
Sbjct 175 GWVWLVLKREERRLEVVKTSNAINPLVWDD--IPIICVDVWEHSYYLDYKNDRAKYINTF 232
Query 202 WS-LVNWDFANENLKKAMA 219
+ LV+W+ A + +A A
Sbjct 233 LNHLVSWNAAMSRMARAEA 251
> At4g25100
Length=212
Score = 166 bits (420), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 97/204 (47%), Positives = 133/204 (65%), Gaps = 10/204 (4%)
Query 23 FQLPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASKSLEDLIKTA-- 80
+ L P P+A DALEPH+SK+TLE+H+GKHH YV+NL + G + K LE +I +
Sbjct 11 YVLKPPPFALDALEPHMSKQTLEFHWGKHHRAYVDNLKKQVLGTELEGKPLEHIIHSTYN 70
Query 81 SGGIL---NNAGQVYNHTFYWNSMKAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAG 137
+G +L NNA Q +NH F+W SMK GGGG P+G + +E+ F+S+E F E+F+AAAA
Sbjct 71 NGDLLPAFNNAAQAWNHEFFWESMKPGGGGKPSGELLALLERDFTSYEKFYEEFNAAAAT 130
Query 138 HFGSGWVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRNDRAAY 197
FG+GW WL Y E+ KVV +T + P S FPLL DVWEHAYY+D +N R Y
Sbjct 131 QFGAGWAWLAYSNEKLKVV--KTPNAVNPLVLGS--FPLLTIDVWEHAYYLDFQNRRPDY 186
Query 198 VKAWWS-LVNWDFANENLKKAMAA 220
+K + + LV+W+ + L+ A AA
Sbjct 187 IKTFMTNLVSWEAVSARLEAAKAA 210
> ECU11g1080
Length=233
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 79/203 (38%), Positives = 105/203 (51%), Gaps = 11/203 (5%)
Query 21 MPFQLPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASKSLEDLIKTA 80
M F+LP L Y DALEP I +ET+ H+ KHH Y+N+L + KSL +
Sbjct 8 MEFKLPELDYPYDALEPIIDEETMRTHHSKHHQAYINSLEKTLRSNSIKGKSLYYYVTKG 67
Query 81 SG----------GILNNAGQVYNHTFYWNSMKAGGGGSPTGLIAGE-IEKSFSSFEAFKE 129
G +LN +G YNH+ +W M G P E IE+SF S E +
Sbjct 68 RGIRGVKSSARRDLLNFSGGHYNHSLFWKMMCPPGTSGPMSPRLLEYIERSFGSQEKMVK 127
Query 130 QFSAAAAGHFGSGWVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYID 189
+FS AA FGSGWVWLCY + +VI +T++ S P+L DVWEHAYY+
Sbjct 128 EFSNAAVSLFGSGWVWLCYRQSEGILVIRKTYNQDAICMKTSSTVPILGLDVWEHAYYLK 187
Query 190 RRNDRAAYVKAWWSLVNWDFANE 212
+N R+ YV WW +VNW +
Sbjct 188 YKNVRSEYVANWWKVVNWGLVSR 210
> Hs10835187
Length=222
Score = 147 bits (372), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 81/210 (38%), Positives = 113/210 (53%), Gaps = 5/210 (2%)
Query 4 SRTLAVSVDSFKSKDKKMPFQLPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLT 63
SR L + S+ K LP LPY ALEPHI+ + ++ H+ KHHA YVNNLN
Sbjct 10 SRQLPPVLGYLGSRQKH---SLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTE 66
Query 64 EGKDEASKSLEDLIKTASGGILN-NAGQVYNHTFYWNSMKAGGGGSPTGLIAGEIEKSFS 122
E EA + + A L N G NH+ +W ++ GGG P G + I+ F
Sbjct 67 EKYQEALAKGDVTAQIALQPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKLDFG 126
Query 123 SFEAFKEQFSAAAAGHFGSGWVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVW 182
SF+ FKE+ +AA+ G GSGW WL + E+ + I + P + + PLL DVW
Sbjct 127 SFDKFKEKLTAASVGVQGSGWGWLGFNKERGHLQIAACPNQD-PLQGTTGLIPLLGIDVW 185
Query 183 EHAYYIDRRNDRAAYVKAWWSLVNWDFANE 212
EHAYY+ +N R Y+KA W+++NW+ E
Sbjct 186 EHAYYLQYKNVRPDYLKAIWNVINWENVTE 215
> At3g10920
Length=231
Score = 138 bits (348), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 76/199 (38%), Positives = 102/199 (51%), Gaps = 6/199 (3%)
Query 23 FQLPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASK--SLEDLIKTA 80
F LP LPY ALEP IS E ++ H+ KHH YV N N E D+A ++K
Sbjct 31 FTLPDLPYDYGALEPAISGEIMQIHHQKHHQAYVTNYNNALEQLDQAVNKGDASTVVKLQ 90
Query 81 SGGILNNAGQVYNHTFYWNSM---KAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAG 137
S N G V NH+ +W ++ GGG P G + I+ F S E ++ SA A
Sbjct 91 SAIKFNGGGHV-NHSIFWKNLAPSSEGGGEPPKGSLGSAIDAHFGSLEGLVKKMSAEGAA 149
Query 138 HFGSGWVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRNDRAAY 197
GSGWVWL E KK+V+ T + PL+ DVWEHAYY+ +N R Y
Sbjct 150 VQGSGWVWLGLDKELKKLVVDTTANQDPLVTKGGSLVPLVGIDVWEHAYYLQYKNVRPEY 209
Query 198 VKAWWSLVNWDFANENLKK 216
+K W ++NW +A+E +K
Sbjct 210 LKNVWKVINWKYASEVYEK 228
> CE09323
Length=221
Score = 135 bits (341), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 76/197 (38%), Positives = 103/197 (52%), Gaps = 9/197 (4%)
Query 25 LPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASK--SLEDLIKTASG 82
LP LPY LEP IS E ++ H+ KHHATYVNNLN++ E EA ++++ I
Sbjct 28 LPDLPYDYADLEPVISHEIMQLHHQKHHATYVNNLNQIEEKLHEAVSKGNVKEAIALQPA 87
Query 83 GILNNAGQVYNHTFYWNSMKAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAGHFGSG 142
N G + NH+ +W ++ A GG P+ + I+ F S + ++Q SA+ GSG
Sbjct 88 LKFNGGGHI-NHSIFWTNL-AKDGGEPSAELLTAIKSDFGSLDNLQKQLSASTVAVQGSG 145
Query 143 WVWLCYC--GEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRNDRAAYVKA 200
W WL YC G+ KV D P + PL DVWEHAYY+ +N R YV A
Sbjct 146 WGWLGYCPKGKILKVATCANQD---PLEATTGLVPLFGIDVWEHAYYLQYKNVRPDYVNA 202
Query 201 WWSLVNWDFANENLKKA 217
W + NW +E KA
Sbjct 203 IWKIANWKNVSERFAKA 219
> At3g56350
Length=241
Score = 134 bits (338), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 75/203 (36%), Positives = 104/203 (51%), Gaps = 18/203 (8%)
Query 25 LPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNR--------LTEGKDEASKSLEDL 76
LP LPYA DALEP IS+E + H+ KHH TYV N+ + +G + L+ L
Sbjct 38 LPDLPYAYDALEPAISEEIMRLHHQKHHQTYVTQYNKALNSLRSAMADGDHSSVVKLQSL 97
Query 77 IKTASGGILNNAGQVYNHTFYWNSMKA---GGGGSPTGLIAGEIEKSFSSFEAFKEQFSA 133
IK N G V NH +W ++ GGG P +A I+ F S E ++ +A
Sbjct 98 IK------FNGGGHV-NHAIFWKNLAPVHEGGGKPPHDPLASAIDAHFGSLEGLIQKMNA 150
Query 134 AAAGHFGSGWVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRND 193
A GSGWVW E K++V+ T + S PL+ DVWEHAYY +N
Sbjct 151 EGAAVQGSGWVWFGLDRELKRLVVETTANQDPLVTKGSHLVPLIGIDVWEHAYYPQYKNA 210
Query 194 RAAYVKAWWSLVNWDFANENLKK 216
RA Y+K W+++NW +A + +K
Sbjct 211 RAEYLKNIWTVINWKYAADVFEK 233
> 7302882
Length=217
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 72/186 (38%), Positives = 100/186 (53%), Gaps = 5/186 (2%)
Query 25 LPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEA-SKS-LEDLIKTASG 82
LP LPY ALEP I +E +E H+ KHH TYVNNLN E +EA SKS LI+ A
Sbjct 21 LPKLPYDYAALEPIICREIMELHHQKHHQTYVNNLNAAEEQLEEAKSKSDTTKLIQLAPA 80
Query 83 GILNNAGQVYNHTFYWNSMKAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAGHFGSG 142
N G + NHT +W ++ + P+ + IE + S E FK++ + GSG
Sbjct 81 LRFNGGGHI-NHTIFWQNL-SPNKTQPSDDLKKAIESQWKSLEEFKKELTTLTVAVQGSG 138
Query 143 WVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRNDRAAYVKAWW 202
W WL + + K+ + + P ++ PL DVWEHAYY+ +N R +YV+A W
Sbjct 139 WGWLGFNKKSGKLQLAALPNQD-PLEASTGLIPLFGIDVWEHAYYLQYKNVRPSYVEAIW 197
Query 203 SLVNWD 208
+ NWD
Sbjct 198 DIANWD 203
> CE08002
Length=218
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 71/195 (36%), Positives = 101/195 (51%), Gaps = 8/195 (4%)
Query 25 LPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASK--SLEDLIKTASG 82
LP LP+ LEP IS E ++ H+ KHHATYVNNLN++ E EA +L++ I
Sbjct 28 LPDLPFDYADLEPVISHEIMQLHHQKHHATYVNNLNQIEEKLHEAVSKGNLKEAIALQPA 87
Query 83 GILNNAGQVYNHTFYWNSMKAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAGHFGSG 142
N G + NH+ +W ++ A GG P+ + I++ F S + +++ S GSG
Sbjct 88 LKFNGGGHI-NHSIFWTNL-AKDGGEPSKELMDTIKRDFGSLDNLQKRLSDITIAVQGSG 145
Query 143 WVWLCYCGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRNDRAAYVKAWW 202
W WL YC ++ K++ T P PL DVWEHAYY+ +N R YV A W
Sbjct 146 WGWLGYC-KKDKILKIATCANQDPLEG---MVPLFGIDVWEHAYYLQYKNVRPDYVHAIW 201
Query 203 SLVNWDFANENLKKA 217
+ NW +E A
Sbjct 202 KIANWKNISERFANA 216
> SPAC1486.01
Length=218
Score = 122 bits (306), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 70/191 (36%), Positives = 101/191 (52%), Gaps = 12/191 (6%)
Query 25 LPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASKSLEDLIKTASGGI 84
LPPLPYA +ALEP +S+ ++ H+ KHH TYVNNLN E + + LE + +
Sbjct 28 LPPLPYAYNALEPALSETIMKLHHDKHHQTYVNNLNAAQEKLADPNLDLEGEVALQAAIK 87
Query 85 LNNAGQVYNHTFYWNSM--KAGGGGSP--TGLIAGEIEKSFSSFEAFKEQFSAAAAGHFG 140
N G + NH+ +W + + GGG P +G + I + S E F+++ +AA A G
Sbjct 88 FNGGGHI-NHSLFWKILAPQKEGGGKPVTSGSLHKAITSKWGSLEDFQKEMNAALASIQG 146
Query 141 SGWVWLCYCGE-QKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAYYIDRRNDRAAYVK 199
SGW WL + ++ T D V K P++ D WEHAYY N +A Y K
Sbjct 147 SGWAWLIVDKDGSLRITTTANQDTIV------KSKPIIGIDAWEHAYYPQYENRKAEYFK 200
Query 200 AWWSLVNWDFA 210
A W+++NW A
Sbjct 201 AIWNVINWKEA 211
> YHR008c
Length=233
Score = 107 bits (266), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 67/201 (33%), Positives = 96/201 (47%), Gaps = 15/201 (7%)
Query 19 KKMPFQLPPLPYAKDALEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASKSLEDLIK 78
++ LP L + ALEP+IS + E HY KHH TYVN N + E S L
Sbjct 24 RRTKVTLPDLKWDFGALEPYISGQINELHYTKHHQTYVNGFNTAVDQFQELSDLLAKEPS 83
Query 79 TASG--------GILNNAGQVYNHTFYWNSM---KAGGGGSPTGLIAGEIEKSFSSFEAF 127
A+ I + G NH +W ++ GGG PTG +A I++ F S +
Sbjct 84 PANARKMIAIQQNIKFHGGGFTNHCLFWENLAPESQGGGEPPTGALAKAIDEQFGSLDEL 143
Query 128 KEQFSAAAAGHFGSGWVWLCY-CGEQKKVVITQTHDGGVPFRDNSKCFPLLACDVWEHAY 186
+ + AG GSGW ++ K+ + QT++ PL+A D WEHAY
Sbjct 144 IKLTNTKLAGVQGSGWAFIVKNLSNGGKLDVVQTYNQDTV---TGPLVPLVAIDAWEHAY 200
Query 187 YIDRRNDRAAYVKAWWSLVNW 207
Y+ +N +A Y KA W++VNW
Sbjct 201 YLQYQNKKADYFKAIWNVVNW 221
> SPBC3H7.04
Length=220
Score = 68.9 bits (167), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 44/152 (28%), Positives = 67/152 (44%), Gaps = 18/152 (11%)
Query 80 ASGGILNNAGQVYNHTFYWNSM------KAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSA 133
A + N + Q+ NH F+++ + A L G I+ SF SF K Q
Sbjct 64 ARANLFNYSSQLVNHDFFFSGLISPERPSADADLGAINLKPG-IDASFGSFGELKSQMVD 122
Query 134 AAAGHFGSGWVWLCYCGEQKKVVITQTHDG--------GVP-FRDNSKCFPLLACDVWEH 184
FG GW+WL Y E+ + T++ G P FR N+ PLL ++W++
Sbjct 123 VGNSVFGDGWLWLVYSPEKSLFSLLCTYNASNAFLWGTGFPKFRTNA-IVPLLCVNLWQY 181
Query 185 AYYIDR-RNDRAAYVKAWWSLVNWDFANENLK 215
AY D N + Y+ WW ++NW N +
Sbjct 182 AYLDDYGLNGKKMYITKWWDMINWTVVNNRFQ 213
> SPBC16A3.14
Length=277
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 49/221 (22%), Positives = 88/221 (39%), Gaps = 37/221 (16%)
Query 35 LEPHISKETLEYHYGKHHATYVNNLNRLTEGKDEASKSLEDLI-KTAS----GGILNNAG 89
L P S E L+ + +H V LN +G + S+ ++I +TA+ A
Sbjct 49 LLPLFSPEALDIAWDQHQRQVVKELNDRVKGTELEDSSVFNIIFQTAALPEHAATFQFAS 108
Query 90 QVYNHTFYWNSM--------KAGGGGSPTGLIAGEIEKSFSSFEAFKEQFSAAAAGHFGS 141
Q YN+ F++ S+ K I + ++F S E + A+ FG+
Sbjct 109 QAYNNHFFFQSLIGKRAADAKKNSKYEANAAINKAVNENFGSKENLLSKIHELASNSFGA 168
Query 142 GWVWLCYCGEQKKVVITQTHDGGVPF-----RDNSKCF-----------------PLLAC 179
W+W+ + ++ + +T G P+ + N P+L
Sbjct 169 CWLWIV-IDDYNRLNLLRTFQAGSPYLWTRWQSNDPHLISSVPDYSARPRKYAHVPILNL 227
Query 180 DVWEHAYYIDR-RNDRAAYVKAWWSLVNWDFANENLKKAMA 219
+W HAYY D +R+ Y+ W+ ++W E L ++A
Sbjct 228 CLWNHAYYKDYGLLNRSRYIDTWFDCIDWSVIEERLTNSLA 268
> YJR101w
Length=266
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 58/253 (22%), Positives = 99/253 (39%), Gaps = 66/253 (26%)
Query 25 LPPLPYAKDALE---PHISK----ETLEYHYGKHHATYVNNLNRLTEGKD-EASKSLEDL 76
+P LP +K L+ P+I +T+ + Y ++ + L T G+ E+ L
Sbjct 11 VPKLPNSKALLQNGVPNILSSSGFKTVWFDYQRY---LCDKLTLATAGQSLESYYPFHIL 67
Query 77 IKTA----SGGILNNAGQVYN-HTFYWNSMKAG---GGGS-------PTGLIAGEIEKSF 121
+KTA I N A ++N H F N + + G S P+ L +I+ SF
Sbjct 68 LKTAGNPLQSNIFNLASSIHNNHLFVENILPSAVEHGTNSNAVVKTEPSRLFLSKIKDSF 127
Query 122 --SSFEAFKEQFSAAAAGH-FGSGWVWLCYCGEQKKVVITQTHDGGVPFRDNSKCF---- 174
S +E KE+ A G GW++L E+K ++T ++G + ++ F
Sbjct 128 NGSDWEVVKEEMIYRAENEVLGQGWLFLVENNEKKLFILTSNNNGTPYYFPRNQSFDLNS 187
Query 175 --------------------------------PLLACDVWEHAYYIDR-RNDRAAYVKAW 201
P++ ++W+HAY D +R+ YVK
Sbjct 188 AISIDEFATLKQMKELIGKSTKLNGKVQDWTMPIICVNLWDHAYLHDYGVGNRSKYVKNV 247
Query 202 WSLVNWDFANENL 214
+NW N +
Sbjct 248 LDNLNWSVVNNRI 260
> CE06331
Length=376
Score = 33.9 bits (76), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 149 CGEQKKVVITQTHDGGVPFRDNSKCFPLLA 178
C QK +VI QT DG VP R + FPL++
Sbjct 282 CRRQKTIVIEQTVDGTVPARRVTFGFPLVS 311
> Hs8922999
Length=509
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 37/83 (44%), Gaps = 17/83 (20%)
Query 30 YAKDALEPHISKETLEYHYGKHHATY---VNNLNRLTEGKDEASKSLEDLIKTASGGILN 86
+A ++ PH+ ++++EY K N+L+ +T+ +EAS EDL+
Sbjct 229 FAISSIRPHLMQQSVEYERKKFQEILERQPNSLDFVTQWLEEAS---EDLM--------- 276
Query 87 NAGQVYNHTFYWNSMKAGGGGSP 109
Q Y H M AG G P
Sbjct 277 --TQKYKHALPVGGMAAGSGDMP 297
> At3g11540
Length=914
Score = 28.9 bits (63), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 25/55 (45%), Gaps = 5/55 (9%)
Query 44 LEYHYGKHHATYVNNLNRLTEGKDEASKSLEDL-----IKTASGGILNNAGQVYN 93
L +H+ H A NNL L + +D K++E IK LNN G VY
Sbjct 320 LAFHFNPHCAEACNNLGVLYKDRDNLDKAVECYQMALSIKPNFAQSLNNLGVVYT 374
> YNL039w
Length=594
Score = 28.9 bits (63), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 2/48 (4%)
Query 50 KHHATYVNNLNRLTEGKDEASKSLED--LIKTASGGILNNAGQVYNHT 95
KHH + + +D+ ++ L D L K SGGI+ N +VY T
Sbjct 510 KHHMKEIEEAKNTAKEEDQTAQRLNDANLNKKGSGGIMTNDLKVYRKT 557
Lambda K H
0.316 0.132 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4147789496
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40