bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0220_orf2
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
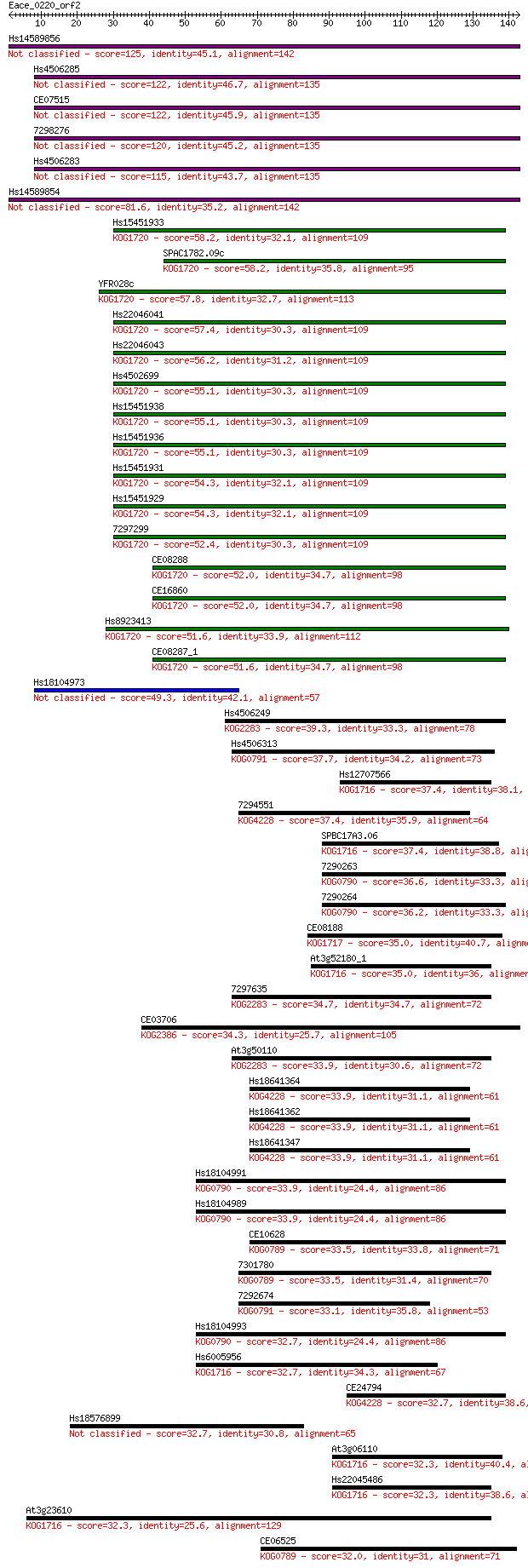
Hs14589856 125 4e-29
Hs4506285 122 2e-28
CE07515 122 3e-28
7298276 120 1e-27
Hs4506283 115 2e-26
Hs14589854 81.6 4e-16
Hs15451933 58.2 5e-09
SPAC1782.09c 58.2 5e-09
YFR028c 57.8 6e-09
Hs22046041 57.4 9e-09
Hs22046043 56.2 2e-08
Hs4502699 55.1 4e-08
Hs15451938 55.1 4e-08
Hs15451936 55.1 4e-08
Hs15451931 54.3 8e-08
Hs15451929 54.3 8e-08
7297299 52.4 3e-07
CE08288 52.0 4e-07
CE16860 52.0 4e-07
Hs8923413 51.6 5e-07
CE08287_1 51.6 5e-07
Hs18104973 49.3 2e-06
Hs4506249 39.3 0.003
Hs4506313 37.7 0.007
Hs12707566 37.4 0.008
7294551 37.4 0.008
SPBC17A3.06 37.4 0.009
7290263 36.6 0.018
7290264 36.2 0.021
CE08188 35.0 0.042
At3g52180_1 35.0 0.047
7297635 34.7 0.066
CE03706 34.3 0.072
At3g50110 33.9 0.094
Hs18641364 33.9 0.10
Hs18641362 33.9 0.10
Hs18641347 33.9 0.10
Hs18104991 33.9 0.11
Hs18104989 33.9 0.11
CE10628 33.5 0.13
7301780 33.5 0.15
7292674 33.1 0.16
Hs18104993 32.7 0.20
Hs6005956 32.7 0.24
CE24794 32.7 0.24
Hs18576899 32.7 0.26
At3g06110 32.3 0.27
Hs22045486 32.3 0.27
At3g23610 32.3 0.30
CE06525 32.0 0.37
> Hs14589856
Length=173
Score = 125 bits (313), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 64/144 (44%), Positives = 88/144 (61%), Gaps = 2/144 (1%)
Query 1 AGMVLNVPTLIESRGVKCLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGA 60
A M P + + ++ LI PTN L ++ L + G T +VR C TYD+ P+
Sbjct 2 ARMNRPAPVEVSYKHMRFLITHNPTNATLSTFIEDLKKYGATTVVRVCEVTYDKTPLEKD 61
Query 61 GIRVHDLTFPDGEAPPAEVIARWRAL--AAQAQAEGGVLAVHCVAGLGRGPVLVAVSLID 118
GI V D F DG PP +V+ W +L A +A G +AVHCVAGLGR PVLVA++LI+
Sbjct 62 GITVVDWPFDDGAPPPGKVVEDWLSLVKAKFCEAPGSCVAVHCVAGLGRAPVLVALALIE 121
Query 119 SGFEAEEAVNFIRSRRKGAINRRQ 142
SG + E+A+ FIR +R+GAIN +Q
Sbjct 122 SGMKYEDAIQFIRQKRRGAINSKQ 145
> Hs4506285
Length=167
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 63/137 (45%), Positives = 81/137 (59%), Gaps = 2/137 (1%)
Query 8 PTLIESRGVKCLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDL 67
P I ++ LI PTN L + +L + GVT LVR C TYD+ PV GI V D
Sbjct 6 PVEISYENMRFLITHNPTNATLNKFTEELKKYGVTTLVRVCDATYDKAPVEKEGIHVLDW 65
Query 68 TFPDGEAPPAEVIARWRALAAQAQAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 125
F DG PP +++ W L E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 66 PFDDGAPPPNQIVDDWLNLLKTKFREEPGCCVAVHCVAGLGRAPVLVALALIECGMKYED 125
Query 126 AVNFIRSRRKGAINRRQ 142
AV FIR +R+GA N +Q
Sbjct 126 AVQFIRQKRRGAFNSKQ 142
> CE07515
Length=190
Score = 122 bits (305), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 62/137 (45%), Positives = 87/137 (63%), Gaps = 2/137 (1%)
Query 8 PTLIESRGVKCLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDL 67
P+ I ++ LI D P N ++Q+Y+ +L + G +VR C PTYD + AGI V D
Sbjct 24 PSEIAWGKMRFLITDRPNNSSIQSYIEELEKHGARAVVRVCEPTYDTLALKEAGIDVLDW 83
Query 68 TFPDGEAPPAEVIARWRALAAQAQAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 125
F DG PP EVI W L + E +AVHCVAGLGR PVLVA++LI++G + E+
Sbjct 84 QFSDGSPPPPEVIKSWFQLCMTSFKEHPDKSIAVHCVAGLGRAPVLVAIALIEAGMKYED 143
Query 126 AVNFIRSRRKGAINRRQ 142
AV IR++R+GA+N++Q
Sbjct 144 AVEMIRTQRRGALNQKQ 160
> 7298276
Length=178
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 61/139 (43%), Positives = 83/139 (59%), Gaps = 4/139 (2%)
Query 8 PTLIESRGVKCLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDL 67
P LIE +G+K LI D P++ + Y+ +L + V +VR C P+Y+ + GI V DL
Sbjct 14 PALIEYKGMKFLITDRPSDITINHYIMELKKNNVNTVVRVCEPSYNTDELETQGITVKDL 73
Query 68 TFPDGEAPPAEVIARWR----ALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEA 123
F DG PP +V+ W L Q +AVHCVAGLGR PVLVA++LI+ G +
Sbjct 74 AFEDGTFPPQQVVDEWFEFFVVLYRYQQNPEACVAVHCVAGLGRAPVLVALALIELGLKY 133
Query 124 EEAVNFIRSRRKGAINRRQ 142
E AV IR +R+GAIN +Q
Sbjct 134 EAAVEMIRDKRRGAINAKQ 152
> Hs4506283
Length=173
Score = 115 bits (289), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 59/137 (43%), Positives = 81/137 (59%), Gaps = 2/137 (1%)
Query 8 PTLIESRGVKCLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDL 67
P + + ++ LI PTN L ++ +L + GVT +VR C TYD V GI V D
Sbjct 9 PVEVTYKNMRFLITHNPTNATLNKFIEELKKYGVTTIVRVCEATYDTTLVEKEGIHVLDW 68
Query 68 TFPDGEAPPAEVIARWRALAAQAQAE--GGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 125
F DG P +++ W +L E G +AVHCVAGLGR PVLVA++LI+ G + E+
Sbjct 69 PFDDGAPPSNQIVDDWLSLVKIKFREEPGCCIAVHCVAGLGRAPVLVALALIEGGMKYED 128
Query 126 AVNFIRSRRKGAINRRQ 142
AV FIR +R+GA N +Q
Sbjct 129 AVQFIRQKRRGAFNSKQ 145
> Hs14589854
Length=148
Score = 81.6 bits (200), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 50/144 (34%), Positives = 68/144 (47%), Gaps = 27/144 (18%)
Query 1 AGMVLNVPTLIESRGVKCLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGA 60
A M P + + ++ LI PTN L ++ L + G T +VR C TYD+ P+
Sbjct 2 ARMNRPAPVEVSYKHMRFLITHNPTNATLSTFIEDLKKYGATTVVRVCEVTYDKTPLEKD 61
Query 61 GIRVHDLTFPDGEAPPAEVIARWRAL--AAQAQAEGGVLAVHCVAGLGRGPVLVAVSLID 118
GI V D F DG PP +V+ W +L A +A G +AVHCVAGLGR
Sbjct 62 GITVVDWPFDDGAPPPGKVVEDWLSLVKAKFCEAPGSCVAVHCVAGLGR----------- 110
Query 119 SGFEAEEAVNFIRSRRKGAINRRQ 142
+R+GAIN +Q
Sbjct 111 --------------KRRGAINSKQ 120
> Hs15451933
Length=383
Score = 58.2 bits (139), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 52/110 (47%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+AY + VT +VR Y+ AG +DL F DG P ++ R+ +
Sbjct 210 EAYFPYFKKHNVTAVVRLNKKIYEAKRFTDAGFEHYDLFFIDGSTPSDNIVRRFLNICEN 269
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDSG-FEAEEAVNFIRSRRKGAI 138
+ G +AVHC AGLGR L+A ++ F E + +IR R G+I
Sbjct 270 TE---GAIAVHCKAGLGRTGTLIACYVMKHYRFTHAEIIAWIRICRPGSI 316
> SPAC1782.09c
Length=537
Score = 58.2 bits (139), Expect = 5e-09, Method: Composition-based stats.
Identities = 34/96 (35%), Positives = 49/96 (51%), Gaps = 2/96 (2%)
Query 44 LVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQAQAEGGVLAVHCVA 103
+VR P YD+ GIR ++ F DG P ++ + L + + E GV+AVHC A
Sbjct 230 IVRLNGPLYDKKTFENVGIRHKEMYFEDGTVPELSLVKEFIDLTEEVE-EDGVIAVHCKA 288
Query 104 GLGRGPVLVAVSLI-DSGFEAEEAVNFIRSRRKGAI 138
GLGR L+ LI F A E + ++R R G +
Sbjct 289 GLGRTGCLIGAYLIYKHCFTANEVIAYMRIMRPGMV 324
> YFR028c
Length=551
Score = 57.8 bits (138), Expect = 6e-09, Method: Composition-based stats.
Identities = 37/114 (32%), Positives = 53/114 (46%), Gaps = 1/114 (0%)
Query 26 NENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRA 85
N+ ++ L V +VR Y++ GI+ DL F DG P ++ +
Sbjct 208 NQPFKSVLNFFANNNVQLVVRLNSHLYNKKHFEDIGIQHLDLIFEDGTCPDLSIVKNFVG 267
Query 86 LAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGAI 138
A GG +AVHC AGLGR L+ LI + GF A E + F+R R G +
Sbjct 268 AAETIIKRGGKIAVHCKAGLGRTGCLIGAHLIYTYGFTANECIGFLRFIRPGMV 321
> Hs22046041
Length=400
Score = 57.4 bits (137), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+ Y+ VT ++R +D AG HDL F DG P ++ R+ +
Sbjct 169 ETYIQYFKNHNVTTIIRLNKRIHDAKRFTDAGFDHHDLFFADGSTPTDAIVKRFLDICEN 228
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDSG-FEAEEAVNFIRSRRKGAI 138
A+ G +AVHC AGLGR L+A ++ A E + ++R R G +
Sbjct 229 AE---GAIAVHCKAGLGRTGTLIACYIMKHYRMTAAETIAWVRICRPGLV 275
> Hs22046043
Length=447
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 34/110 (30%), Positives = 51/110 (46%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+ Y+ VT ++R YD AG HDL F DG P ++ R+ +
Sbjct 216 ETYIQYFKNHNVTTIIRLNKRMYDAKRFTDAGFDHHDLFFADGSTPTDAIVKRFLDICEN 275
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGAI 138
A+ G +AVHC AGLGR L+A ++ A E + ++R R G +
Sbjct 276 AE---GAIAVHCKAGLGRTGTLIACYIMKHYRMTAAETIAWVRICRPGLV 322
> Hs4502699
Length=459
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+ Y+ VT ++R YD AG HDL F DG P ++ + +
Sbjct 246 ETYIQYFKNHNVTTIIRLNKRMYDAKRFTDAGFDHHDLFFADGSTPTDAIVKEFLDICEN 305
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGAI 138
A+ G +AVHC AGLGR L+A ++ A E + ++R R G++
Sbjct 306 AE---GAIAVHCKAGLGRTGTLIACYIMKHYRMTAAETIAWVRICRPGSV 352
> Hs15451938
Length=471
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+ Y+ VT ++R YD AG HDL F DG P ++ + +
Sbjct 246 ETYIQYFKNHNVTTIIRLNKRMYDAKRFTDAGFDHHDLFFADGSTPTDAIVKEFLDICEN 305
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGAI 138
A+ G +AVHC AGLGR L+A ++ A E + ++R R G++
Sbjct 306 AE---GAIAVHCKAGLGRTGTLIACYIMKHYRMTAAETIAWVRICRPGSV 352
> Hs15451936
Length=498
Score = 55.1 bits (131), Expect = 4e-08, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+ Y+ VT ++R YD AG HDL F DG P ++ + +
Sbjct 246 ETYIQYFKNHNVTTIIRLNKRMYDAKRFTDAGFDHHDLFFADGSTPTDAIVKEFLDICEN 305
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGAI 138
A+ G +AVHC AGLGR L+A ++ A E + ++R R G++
Sbjct 306 AE---GAIAVHCKAGLGRTGTLIACYIMKHYRMTAAETIAWVRICRPGSV 352
> Hs15451931
Length=623
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 35/110 (31%), Positives = 52/110 (47%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+AY + VT +VR Y+ AG +DL F DG P ++ R+ +
Sbjct 210 EAYFPYFKKHNVTAVVRLNKKIYEAKRFTDAGFEHYDLFFIDGSTPSDNIVRRFLNICEN 269
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGAI 138
+ G +AVHC AGLGR L+A ++ F E + +IR R G+I
Sbjct 270 TE---GAIAVHCKAGLGRTGTLIACYVMKHYRFTHAEIIAWIRICRPGSI 316
> Hs15451929
Length=594
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 35/110 (31%), Positives = 52/110 (47%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+AY + VT +VR Y+ AG +DL F DG P ++ R+ +
Sbjct 210 EAYFPYFKKHNVTAVVRLNKKIYEAKRFTDAGFEHYDLFFIDGSTPSDNIVRRFLNICEN 269
Query 90 AQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGAI 138
+ G +AVHC AGLGR L+A ++ F E + +IR R G+I
Sbjct 270 TE---GAIAVHCKAGLGRTGTLIACYVMKHYRFTHAEIIAWIRICRPGSI 316
> 7297299
Length=553
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 4/110 (3%)
Query 30 QAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQ 89
+ Y + VT ++R Y AG DL F DG P ++ ++ ++
Sbjct 215 ERYFSYFRDNNVTTVIRLNAKVYHASSFENAGFDHKDLFFIDGSTPSDAIMKKFLSICET 274
Query 90 AQAEGGVLAVHCVAGLGR-GPVLVAVSLIDSGFEAEEAVNFIRSRRKGAI 138
+ G +AVHC AGLGR G ++ A + GF A EA+ ++R R G++
Sbjct 275 TK---GAIAVHCKAGLGRTGSLIGAYIMKHYGFTALEAIAWLRLCRPGSV 321
> CE08288
Length=681
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 34/99 (34%), Positives = 51/99 (51%), Gaps = 4/99 (4%)
Query 41 VTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQAQAEGGVLAVH 100
V+ +VR YD AG DL F DG P E++ ++ + +GGV AVH
Sbjct 238 VSTIVRLNAKNYDASKFTKAGFDHVDLFFIDGSTPSDEIMLKF--IKVVDNTKGGV-AVH 294
Query 101 CVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIRSRRKGAI 138
C AGLGR L+A ++ + G A E + ++R R G++
Sbjct 295 CKAGLGRTGTLIACWMMKEYGLTAGECMGWLRVCRPGSV 333
> CE16860
Length=708
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 34/99 (34%), Positives = 51/99 (51%), Gaps = 4/99 (4%)
Query 41 VTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQAQAEGGVLAVH 100
V+ +VR YD AG DL F DG P E++ ++ + +GGV AVH
Sbjct 238 VSTIVRLNAKNYDASKFTKAGFDHVDLFFIDGSTPSDEIMLKF--IKVVDNTKGGV-AVH 294
Query 101 CVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIRSRRKGAI 138
C AGLGR L+A ++ + G A E + ++R R G++
Sbjct 295 CKAGLGRTGTLIACWMMKEYGLTAGECMGWLRVCRPGSV 333
> Hs8923413
Length=150
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 54/116 (46%), Gaps = 8/116 (6%)
Query 28 NLQAYLAQLVQVGVTDLVRTC---PPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWR 84
L A+ L+ +GV LV PP D P G+ +H L PD P + I R+
Sbjct 23 RLPAHYQFLLDLGVRHLVSLTERGPPHSDSCP----GLTLHRLRIPDFCPPAPDQIDRFV 78
Query 85 ALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIRSRRKGAIN 139
+ +A A G + VHC G GR ++A L+ + G A +A+ IR R G I
Sbjct 79 QIVDEANARGEAVGVHCALGFGRTGTMLACYLVKERGLAAGDAIAEIRRLRPGPIE 134
> CE08287_1
Length=753
Score = 51.6 bits (122), Expect = 5e-07, Method: Composition-based stats.
Identities = 34/99 (34%), Positives = 51/99 (51%), Gaps = 4/99 (4%)
Query 41 VTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAEVIARWRALAAQAQAEGGVLAVH 100
V+ +VR YD AG DL F DG P E++ ++ + +GGV AVH
Sbjct 238 VSTIVRLNAKNYDASKFTKAGFDHVDLFFIDGSTPSDEIMLKF--IKVVDNTKGGV-AVH 294
Query 101 CVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIRSRRKGAI 138
C AGLGR L+A ++ + G A E + ++R R G++
Sbjct 295 CKAGLGRTGTLIACWMMKEYGLTAGECMGWLRVCRPGSV 333
> Hs18104973
Length=82
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 24/57 (42%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 8 PTLIESRGVKCLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRV 64
P I ++ LI PTN L + +L + GVT LVR C TYD+ PV GI V
Sbjct 6 PVEISYENMRFLITHNPTNATLNKFTEELKKYGVTTLVRVCDATYDKAPVEKEGIHV 62
> Hs4506249
Length=403
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 42/85 (49%), Gaps = 7/85 (8%)
Query 61 GIRVHDLTFPDGEAPPAEVIARWRALAAQ--AQAEGGVLAVHCVAGLGRGPVLVAVSLID 118
RV F D P E+I + Q ++ + V A+HC AG GR V++ L+
Sbjct 82 NCRVAQYPFEDHNPPQLELIKPFCEDLDQWLSEDDNHVAAIHCKAGKGRTGVMICAYLLH 141
Query 119 SG--FEAEEAVNF---IRSRRKGAI 138
G +A+EA++F +R+R K +
Sbjct 142 RGKFLKAQEALDFYGEVRTRDKKGV 166
> Hs4506313
Length=1118
Score = 37.7 bits (86), Expect = 0.007, Method: Composition-based stats.
Identities = 25/80 (31%), Positives = 40/80 (50%), Gaps = 7/80 (8%)
Query 63 RVHDLTFPDGEAP--PAEVIARWRALAA--QAQAEGGVLAVHCVAGLGRGPVLVAVSLID 118
+ H +PD P P ++A WR L EGG VHC AG+GR L+A+ ++
Sbjct 980 QFHYQAWPDHGVPSSPDTLLAFWRMLRQWLDQTMEGGPPIVHCSAGVGRTGTLIALDVLL 1039
Query 119 SGFEAEEAV---NFIRSRRK 135
++E + +F+R R+
Sbjct 1040 RQLQSEGLLGPFSFVRKMRE 1059
> Hs12707566
Length=384
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 1/43 (2%)
Query 93 EGGVLAVHCVAGLGRGPVLVAVSLIDSG-FEAEEAVNFIRSRR 134
+GG + VHC AG+ R P + L+ + F +EA ++I+ RR
Sbjct 255 KGGKVLVHCEAGISRSPTICMAYLMKTKQFRLKEAFDYIKQRR 297
> 7294551
Length=1428
Score = 37.4 bits (85), Expect = 0.008, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 65 HDLTFPDGEAP--PAEVIARWRALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFE 122
H LT+ D AP P +I R + + + G + VHC AG+GR LVA+ + E
Sbjct 1025 HYLTWKDFMAPEHPHGIIKFIRQINSVYSLQRGPILVHCSAGVGRTGTLVALDSLIQQLE 1084
Query 123 AEEAVN 128
E++V+
Sbjct 1085 EEDSVS 1090
> SPBC17A3.06
Length=330
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Query 88 AQAQAEGGVLAVHCVAGLGRGPVLVAVSLI-DSGFEAEEAVNFIRSRRKG 136
A A ++ + VHC AG+ R LVA L+ ++ + EEA++ I RR G
Sbjct 118 AFALSKNAKVLVHCFAGISRSVTLVAAYLMKENNWNTEEALSHINERRSG 167
> 7290263
Length=845
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query 88 AQAQAEGGVLAVHCVAGLGRGPVLVAVSLI---------DSGFEAEEAVNFIRSRRKGAI 138
AQA + G + VHC AG+GR + + +I D+ + + + +RS+R G +
Sbjct 570 AQAGEKPGPICVHCSAGIGRTGTFIVIDMILDQIVRNGLDTEIDIQRTIQMVRSQRSGLV 629
> 7290264
Length=945
Score = 36.2 bits (82), Expect = 0.021, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query 88 AQAQAEGGVLAVHCVAGLGRGPVLVAVSLI---------DSGFEAEEAVNFIRSRRKGAI 138
AQA + G + VHC AG+GR + + +I D+ + + + +RS+R G +
Sbjct 670 AQAGEKPGPICVHCSAGIGRTGTFIVIDMILDQIVRNGLDTEIDIQRTIQMVRSQRSGLV 729
> CE08188
Length=221
Score = 35.0 bits (79), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 27/55 (49%), Gaps = 2/55 (3%)
Query 84 RALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDS-GFEAEEAVNFIRSRRKGA 137
R + E GVL VHC G+ R LVA LI + +AV+FI RR A
Sbjct 116 RIIHDSRSKEEGVL-VHCFLGVSRSATLVAFYLISALSINWRDAVDFIHHRRFSA 169
> At3g52180_1
Length=248
Score = 35.0 bits (79), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 85 ALAAQAQAEGGVLAVHCVAGLGRGP-VLVAVSLIDSGFEAEEAVNFIRSRR 134
L + GGV VHC AG+GR P V + G++ EA + S+R
Sbjct 182 TLYKAVKRNGGVTYVHCTAGMGRAPAVALTYMFWVQGYKLMEAHKLLMSKR 232
> 7297635
Length=514
Score = 34.7 bits (78), Expect = 0.066, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 37/76 (48%), Gaps = 4/76 (5%)
Query 63 RVHDLTFPDGEAPPAEVIARWRALAAQAQAE--GGVLAVHCVAGLGRGPVLVAVSLIDSG 120
RV F D P E+I R+ + E V+AVHC AG GR ++ L+ SG
Sbjct 92 RVAVYPFDDHNPPTIELIQRFCSDVDMWLKEDSSNVVAVHCKAGKGRTGTMICAYLVFSG 151
Query 121 FE--AEEAVNFIRSRR 134
+ A+EA+ + +R
Sbjct 152 IKKSADEALAWYDEKR 167
> CE03706
Length=261
Score = 34.3 bits (77), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 43/112 (38%), Gaps = 8/112 (7%)
Query 38 QVG-VTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPAE-----VIARWRALAAQAQ 91
Q+G V DL T Y + G++ L P E E I + +
Sbjct 110 QIGLVIDLTNT-DRYYKKTEWADHGVKYLKLNCPGHEVNEREDLVQDFINAVKEFVNDKE 168
Query 92 AEGGVLAVHCVAGLGRGPVLVAVSLID-SGFEAEEAVNFIRSRRKGAINRRQ 142
+G ++ VHC GL R L+ +ID + A +A++ R + R
Sbjct 169 NDGKLIGVHCTHGLNRTGYLICRYMIDVDNYSASDAISMFEYYRGHPMEREH 220
> At3g50110
Length=628
Score = 33.9 bits (76), Expect = 0.094, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 36/76 (47%), Gaps = 4/76 (5%)
Query 63 RVHDLTFPDGEAPPAEVIARWRALAAQAQAEG--GVLAVHCVAGLGRGPVLVAVSLIDSG 120
+V F D PP ++I + A E V+ VHC AG+ R +++ L+
Sbjct 267 KVASFPFDDHNCPPIQLIPSFCQSAYTWLKEDIQNVVVVHCKAGMARTGLMICCLLLYLK 326
Query 121 F--EAEEAVNFIRSRR 134
F AEEA+++ +R
Sbjct 327 FFPTAEEAIDYYNQKR 342
> Hs18641364
Length=1256
Score = 33.9 bits (76), Expect = 0.10, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query 68 TFPDGEAP--PAEVIARWRALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 125
++PD P P ++ R + A + G + VHC AG+GR + + + G EAE
Sbjct 768 SWPDHGVPEDPHLLLKLRRRVNAFSNFFSGPIVVHCSAGVGRTGTYIGIDAMLEGLEAEN 827
Query 126 AVN 128
V+
Sbjct 828 KVD 830
> Hs18641362
Length=1143
Score = 33.9 bits (76), Expect = 0.10, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query 68 TFPDGEAP--PAEVIARWRALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 125
++PD P P ++ R + A + G + VHC AG+GR + + + G EAE
Sbjct 655 SWPDHGVPEDPHLLLKLRRRVNAFSNFFSGPIVVHCSAGVGRTGTYIGIDAMLEGLEAEN 714
Query 126 AVN 128
V+
Sbjct 715 KVD 717
> Hs18641347
Length=1304
Score = 33.9 bits (76), Expect = 0.10, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query 68 TFPDGEAP--PAEVIARWRALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFEAEE 125
++PD P P ++ R + A + G + VHC AG+GR + + + G EAE
Sbjct 816 SWPDHGVPEDPHLLLKLRRRVNAFSNFFSGPIVVHCSAGVGRTGTYIGIDAMLEGLEAEN 875
Query 126 AVN 128
V+
Sbjct 876 KVD 878
> Hs18104991
Length=597
Score = 33.9 bits (76), Expect = 0.11, Method: Composition-based stats.
Identities = 21/99 (21%), Positives = 45/99 (45%), Gaps = 13/99 (13%)
Query 53 DEGPVIGAGIRVHDLTFPDGEAP--PAEVIARWRALAAQAQA--EGGVLAVHCVAGLGRG 108
D G +I L++PD P P V++ + + ++ G + VHC AG+GR
Sbjct 403 DNGDLIREIWHYQYLSWPDHGVPSEPGGVLSFLDQINQRQESLPHAGPIIVHCSAGIGRT 462
Query 109 PVLVAVSL---------IDSGFEAEEAVNFIRSRRKGAI 138
++ + + +D + ++ + +R++R G +
Sbjct 463 GTIIVIDMLMENISTKGLDCDIDIQKTIQMVRAQRSGMV 501
> Hs18104989
Length=595
Score = 33.9 bits (76), Expect = 0.11, Method: Composition-based stats.
Identities = 21/99 (21%), Positives = 45/99 (45%), Gaps = 13/99 (13%)
Query 53 DEGPVIGAGIRVHDLTFPDGEAP--PAEVIARWRALAAQAQA--EGGVLAVHCVAGLGRG 108
D G +I L++PD P P V++ + + ++ G + VHC AG+GR
Sbjct 401 DNGDLIREIWHYQYLSWPDHGVPSEPGGVLSFLDQINQRQESLPHAGPIIVHCSAGIGRT 460
Query 109 PVLVAVSL---------IDSGFEAEEAVNFIRSRRKGAI 138
++ + + +D + ++ + +R++R G +
Sbjct 461 GTIIVIDMLMENISTKGLDCDIDIQKTIQMVRAQRSGMV 499
> CE10628
Length=374
Score = 33.5 bits (75), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 36/78 (46%), Gaps = 9/78 (11%)
Query 68 TFPDGEAPPAEVIARWRALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDSGF------ 121
+PD APP + A L + + VHC AG+GR +V +SLI
Sbjct 234 NWPDHGAPPINMGAI--NLIEAVNYDTNPVVVHCSAGVGRSGTIVGISLIMDKMIQGINC 291
Query 122 -EAEEAVNFIRSRRKGAI 138
+ ++ V IR++R AI
Sbjct 292 KDMKKLVEEIRNQRHYAI 309
> 7301780
Length=1285
Score = 33.5 bits (75), Expect = 0.15, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 5/75 (6%)
Query 65 HDLTFPDGEAP--PAEVIARWRALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLIDSGFE 122
H +PD P P V+ + +A AE G + VHC AG+GR + + + +
Sbjct 632 HYTNWPDHGTPDHPLPVLNFVKKSSAANPAEAGPIVVHCSAGVGRTGTYIVLDAMLKQIQ 691
Query 123 AEEAVN---FIRSRR 134
+ VN F+R R
Sbjct 692 QKNIVNVFGFLRHIR 706
> 7292674
Length=1647
Score = 33.1 bits (74), Expect = 0.16, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 2/55 (3%)
Query 65 HDLTFPDGEAP-PAEVIARW-RALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLI 117
H T+PD P P + + R+ RA + AE + VHC AG+GR + + I
Sbjct 1446 HFTTWPDFGVPNPPQTLVRFVRAFRDRIGAEQRPIVVHCSAGVGRSGTFITLDRI 1500
> Hs18104993
Length=624
Score = 32.7 bits (73), Expect = 0.20, Method: Composition-based stats.
Identities = 21/99 (21%), Positives = 45/99 (45%), Gaps = 13/99 (13%)
Query 53 DEGPVIGAGIRVHDLTFPDGEAP--PAEVIARWRALAAQAQA--EGGVLAVHCVAGLGRG 108
D G +I L++PD P P V++ + + ++ G + VHC AG+GR
Sbjct 401 DNGDLIREIWHYQYLSWPDHGVPSEPGGVLSFLDQINQRQESLPHAGPIIVHCSAGIGRT 460
Query 109 PVLVAVSL---------IDSGFEAEEAVNFIRSRRKGAI 138
++ + + +D + ++ + +R++R G +
Sbjct 461 GTIIVIDMLMENISTKGLDCDIDIQKTIQMVRAQRSGMV 499
> Hs6005956
Length=340
Score = 32.7 bits (73), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 6/73 (8%)
Query 53 DEGPVIGAGIRVHDLT---FPDGEAPPAEVIA---RWRALAAQAQAEGGVLAVHCVAGLG 106
E P AG V DL P + P ++++ R A QA+AEG + VHC AG+
Sbjct 61 SEEPSFKAGPGVEDLWRLFVPALDKPETDLLSHLDRCVAFIGQARAEGRAVLVHCHAGVS 120
Query 107 RGPVLVAVSLIDS 119
R ++ L+ +
Sbjct 121 RSVAIITAFLMKT 133
> CE24794
Length=2234
Score = 32.7 bits (73), Expect = 0.24, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 6/50 (12%)
Query 95 GVLAVHCVAGLGRGPVLVAVSLIDSGFEAEEAVN------FIRSRRKGAI 138
G + VHC +G GR V +A+S+I AE V+ +R+ R+ I
Sbjct 2161 GPITVHCCSGAGRTAVFIALSIILDRMRAEHVVDVFTTVKLLRTERQNMI 2210
> Hs18576899
Length=96
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 27/65 (41%), Gaps = 17/65 (26%)
Query 18 CLILDAPTNENLQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRVHDLTFPDGEAPPA 77
C +LDAP+ L C P D GP++ IR+ +LT P G P
Sbjct 44 CSLLDAPSTVRL-----------------LCSPGKDNGPLLSDAIRLLELTVPSGHWYPP 86
Query 78 EVIAR 82
E + R
Sbjct 87 EGLVR 91
> At3g06110
Length=167
Score = 32.3 bits (72), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 27/48 (56%), Gaps = 2/48 (4%)
Query 91 QAEGGVLAVHCVAGLGRG-PVLVAVSLIDSGFEAEEAVNFIRSRRKGA 137
Q+ GGVL VHC G+ R ++VA + G +A+ +RSRR A
Sbjct 100 QSGGGVL-VHCFMGMSRSVTIVVAYLMKKHGMGFSKAMELVRSRRHQA 146
> Hs22045486
Length=234
Score = 32.3 bits (72), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 91 QAEGGVLAVHCVAGLGRGPVLVAVSLIDSG-FEAEEAVNFIRSRR 134
+ +G VL VHC AG P + SLI + F +EA N+++ +R
Sbjct 176 EKKGKVL-VHCEAGFSCSPTICMASLIKTKHFCLKEAFNYVKWKR 219
> At3g23610
Length=198
Score = 32.3 bits (72), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 60/131 (45%), Gaps = 12/131 (9%)
Query 6 NVPTLIESRGVKCLILDAPTNEN-LQAYLAQLVQVGVTDLVRTCPPTYDEGPVIGAGIRV 64
NVP+LIE +G+ + A +N+N L++Y + + L P + + +RV
Sbjct 49 NVPSLIE-QGLYLGSVAAASNKNVLKSYNVTHILTVASSLRPAHPDDF-----VYKVVRV 102
Query 65 HDLTFPDGEAPPAEVIARWRALAAQAQAEGGVLAVHCVAGLGRG-PVLVAVSLIDSGFEA 123
D + E E + +A+ +GG + VHC G R ++VA + G
Sbjct 103 VDKEDTNLEMYFDECVD----FIDEAKRQGGSVLVHCFVGKSRSVTIVVAYLMKKHGMTL 158
Query 124 EEAVNFIRSRR 134
+A+ ++S+R
Sbjct 159 AQALQHVKSKR 169
> CE06525
Length=490
Score = 32.0 bits (71), Expect = 0.37, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 40/79 (50%), Gaps = 10/79 (12%)
Query 71 DGEAP--PAEVIARWRALAAQAQAEGGVLAVHCVAGLGRGPVLVAVSLI------DSGFE 122
D AP PA +I ++A+ ++ ++ + VHC AG+GR V + L +S +
Sbjct 224 DQVAPLDPAPMIKMYKAVLNKSSSKP--IVVHCSAGVGRTSTFVGIYLSQIMIRENSSVD 281
Query 123 AEEAVNFIRSRRKGAINRR 141
E + +R+ R GA+ +
Sbjct 282 MVEILKRLRTMRLGAVQSQ 300
Lambda K H
0.320 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40