bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0250_orf3
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
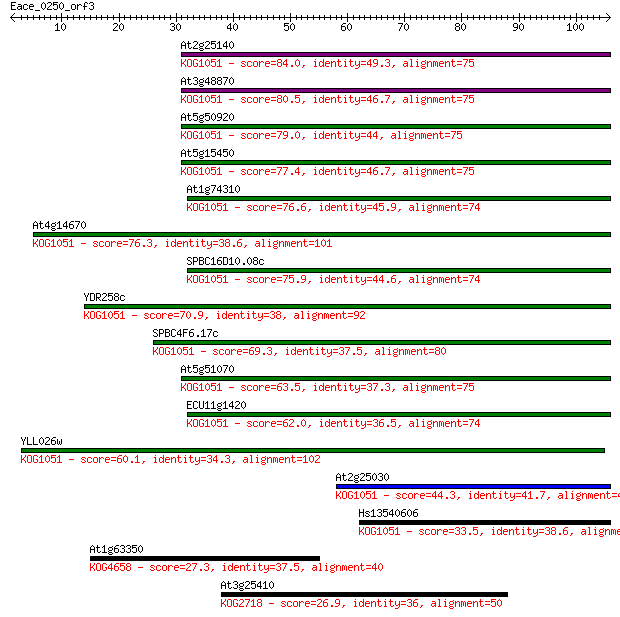
At2g25140 84.0 7e-17
At3g48870 80.5 7e-16
At5g50920 79.0 2e-15
At5g15450 77.4 5e-15
At1g74310 76.6 1e-14
At4g14670 76.3 1e-14
SPBC16D10.08c 75.9 2e-14
YDR258c 70.9 5e-13
SPBC4F6.17c 69.3 1e-12
At5g51070 63.5 1e-10
ECU11g1420 62.0 2e-10
YLL026w 60.1 1e-09
At2g25030 44.3 5e-05
Hs13540606 33.5 0.10
At1g63350 27.3 7.6
At3g25410 26.9 8.0
> At2g25140
Length=874
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 37/75 (49%), Positives = 53/75 (70%), Gaps = 0/75 (0%)
Query 31 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 90
D+A I+ WTGIPL + + E +++ L ++L RVIGQ AVK+VADA+ RAGLS
Sbjct 530 DIAEIVSKWTGIPLSNLQQSEREKLVMLEEVLHHRVIGQDMAVKSVADAIRRSRAGLSDP 589
Query 91 NKPLGTFMFLGSSGV 105
N+P+ +FMF+G +GV
Sbjct 590 NRPIASFMFMGPTGV 604
> At3g48870
Length=952
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 35/75 (46%), Positives = 53/75 (70%), Gaps = 0/75 (0%)
Query 31 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 90
D+ HI+ WTGIP+ K++ DE SR+L++ L +RVIGQ +AVKA++ A+ R GL
Sbjct 596 DIQHIVATWTGIPVEKVSSDESSRLLQMEQTLHTRVIGQDEAVKAISRAIRRARVGLKNP 655
Query 91 NKPLGTFMFLGSSGV 105
N+P+ +F+F G +GV
Sbjct 656 NRPIASFIFSGPTGV 670
> At5g50920
Length=929
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 33/75 (44%), Positives = 52/75 (69%), Gaps = 0/75 (0%)
Query 31 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 90
D+ HI+ WTGIP+ K++ DE R+L++ + L R+IGQ +AVKA++ A+ R GL
Sbjct 575 DIQHIVSSWTGIPVEKVSTDESDRLLKMEETLHKRIIGQDEAVKAISRAIRRARVGLKNP 634
Query 91 NKPLGTFMFLGSSGV 105
N+P+ +F+F G +GV
Sbjct 635 NRPIASFIFSGPTGV 649
> At5g15450
Length=968
Score = 77.4 bits (189), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 35/75 (46%), Positives = 51/75 (68%), Gaps = 0/75 (0%)
Query 31 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 90
D+A I+ WTGIP+ K+ + E ++L L + L RV+GQ AV AVA+A+ RAGLS
Sbjct 615 DIAEIVSKWTGIPVSKLQQSERDKLLHLEEELHKRVVGQNPAVTAVAEAIQRSRAGLSDP 674
Query 91 NKPLGTFMFLGSSGV 105
+P+ +FMF+G +GV
Sbjct 675 GRPIASFMFMGPTGV 689
> At1g74310
Length=911
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 34/74 (45%), Positives = 52/74 (70%), Gaps = 0/74 (0%)
Query 32 VAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKN 91
+A ++ WTGIP+ ++ ++E R++ LAD L RV+GQ QAV AV++A+ RAGL
Sbjct 537 IAEVVSRWTGIPVTRLGQNEKERLIGLADRLHKRVVGQNQAVNAVSEAILRSRAGLGRPQ 596
Query 92 KPLGTFMFLGSSGV 105
+P G+F+FLG +GV
Sbjct 597 QPTGSFLFLGPTGV 610
> At4g14670
Length=623
Score = 76.3 bits (186), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 65/103 (63%), Gaps = 2/103 (1%)
Query 5 LQKWKDELAALEELTANGRRLALSL--HDVAHILHLWTGIPLGKMTEDEISRVLRLADIL 62
+Q+ + +A LE+ + L ++ ++A ++ WTGIP+ ++ ++E R++ LAD L
Sbjct 473 IQEVESAIAKLEKSAKDNVMLTETVGPENIAEVVSRWTGIPVTRLDQNEKKRLISLADKL 532
Query 63 SSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGV 105
RV+GQ +AVKAVA A+ R GL +P G+F+FLG +GV
Sbjct 533 HERVVGQDEAVKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGV 575
> SPBC16D10.08c
Length=905
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 50/74 (67%), Gaps = 0/74 (0%)
Query 32 VAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKN 91
+ I+ WTGIP+ ++ E R+L + +LS +VIGQ +AV AVA+A+ + RAGLS N
Sbjct 552 INEIVARWTGIPVTRLKTTEKERLLNMEKVLSKQVIGQNEAVTAVANAIRLSRAGLSDPN 611
Query 92 KPLGTFMFLGSSGV 105
+P+ +F+F G SG
Sbjct 612 QPIASFLFCGPSGT 625
> YDR258c
Length=811
Score = 70.9 bits (172), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 62/98 (63%), Gaps = 7/98 (7%)
Query 14 ALEELTANGRRLALSLHD------VAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVI 67
AL E + +G ++ L LHD ++ ++ TGIP + + + R+L + + L RV+
Sbjct 449 ALSEKSKDGDKVNL-LHDSVTSDDISKVVAKMTGIPTETVMKGDKDRLLYMENSLKERVV 507
Query 68 GQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGV 105
GQ +A+ A++DA+ +QRAGL+ + +P+ +FMFLG +G
Sbjct 508 GQDEAIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGT 545
> SPBC4F6.17c
Length=803
Score = 69.3 bits (168), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 52/80 (65%), Gaps = 0/80 (0%)
Query 26 ALSLHDVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRA 85
+++ D+A ++ TGIP + E ++L + + ++IGQ +A+KA+ADA+ + RA
Sbjct 464 SVTSDDIAVVVSRATGIPTTNLMRGERDKLLNMEQTIGKKIIGQDEALKAIADAVRLSRA 523
Query 86 GLSPKNKPLGTFMFLGSSGV 105
GL N+PL +F+FLG +GV
Sbjct 524 GLQNTNRPLASFLFLGPTGV 543
> At5g51070
Length=945
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 28/75 (37%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 31 DVAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPK 90
D+A + +W+GIP+ ++T DE ++ L D L RV+GQ +AV A++ A+ R GL
Sbjct 594 DIAAVASVWSGIPVQQITADERMLLMSLEDQLRGRVVGQDEAVAAISRAVKRSRVGLKDP 653
Query 91 NKPLGTFMFLGSSGV 105
++P+ +F G +GV
Sbjct 654 DRPIAAMLFCGPTGV 668
> ECU11g1420
Length=851
Score = 62.0 bits (149), Expect = 2e-10, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 46/74 (62%), Gaps = 0/74 (0%)
Query 32 VAHILHLWTGIPLGKMTEDEISRVLRLADILSSRVIGQQQAVKAVADALAIQRAGLSPKN 91
VA I+ WTGI + ++T E R++ ++ + R+ GQ AV A+ D++ R GL +
Sbjct 520 VAEIISRWTGIDVKRLTIKENERLMEMSSRIKKRIFGQDHAVDAIVDSILQSRVGLDDDD 579
Query 92 KPLGTFMFLGSSGV 105
+P+G+F+ LG +GV
Sbjct 580 RPVGSFLLLGPTGV 593
> YLL026w
Length=908
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 65/105 (61%), Gaps = 5/105 (4%)
Query 3 EELQKWKDELAALEELTANGRRLALSLHD---VAHILHLWTGIPLGKMTEDEISRVLRLA 59
++++K +D++A EE A + ++ D ++ TGIP+ K++E E +++ +
Sbjct 515 KQIEKLEDQVAE-EERRAGANSMIQNVVDSDTISETAARLTGIPVKKLSESENEKLIHME 573
Query 60 DILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSG 104
LSS V+GQ A+KAV++A+ + R+GL+ +P +F+FLG SG
Sbjct 574 RDLSSEVVGQMDAIKAVSNAVRLSRSGLANPRQP-ASFLFLGLSG 617
> At2g25030
Length=265
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 58 LADILSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGV 105
L IL R+I Q V++VADA+ +AG+S N+ + +FMF+G V
Sbjct 2 LEQILHERIIAQDLDVESVADAIRCSKAGISDPNRLIASFMFMGQPSV 49
> Hs13540606
Length=707
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query 62 LSSRVIGQQQAVKAVADALAIQRAGLSPKNKPLGTFMFLGSSGV 105
L +IGQ+ A+ V A+ + G + PL F+FLGSSG+
Sbjct 343 LKEHIIGQESAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGI 385
> At1g63350
Length=898
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Query 15 LEELTANGRRLALSLHDVAHILHLWTGIPLGKMTEDEISR 54
+EE+ +G + + LHDV + LW LGK E I R
Sbjct 462 MEEVELDGANI-VCLHDVVREMALWIASDLGKQNEAFIVR 500
> At3g25410
Length=394
Score = 26.9 bits (58), Expect = 8.0, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 9/59 (15%)
Query 38 LWTGIPLGKMTEDEISRVLR---------LADILSSRVIGQQQAVKAVADALAIQRAGL 87
L G PL ++E+SR + LA +L+S+ +G QAV A + + GL
Sbjct 306 LCIGSPLSINRKEEVSRTISLCTGMQSSTLAGLLASQFLGSSQAVPAACSVVVMAIMGL 364
Lambda K H
0.318 0.133 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1170944580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40