bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0262_orf1
Length=198
Score E
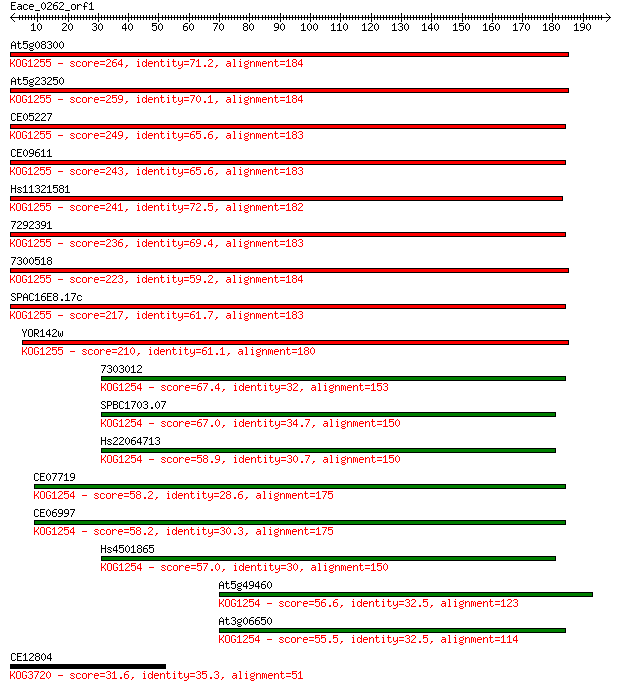
Sequences producing significant alignments: (Bits) Value
At5g08300 264 1e-70
At5g23250 259 3e-69
CE05227 249 2e-66
CE09611 243 2e-64
Hs11321581 241 1e-63
7292391 236 2e-62
7300518 223 2e-58
SPAC16E8.17c 217 1e-56
YOR142w 210 1e-54
7303012 67.4 2e-11
SPBC1703.07 67.0 2e-11
Hs22064713 58.9 6e-09
CE07719 58.2 9e-09
CE06997 58.2 1e-08
Hs4501865 57.0 3e-08
At5g49460 56.6 3e-08
At3g06650 55.5 6e-08
CE12804 31.6 1.1
> At5g08300
Length=347
Score = 264 bits (674), Expect = 1e-70, Method: Compositional matrix adjust.
Identities = 131/184 (71%), Positives = 144/184 (78%), Gaps = 6/184 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
QA+ YGT++V GV P KGGT LPVF SV EAK T +AS I+VP AAA
Sbjct 76 QAIEYGTKMVAGVTPKKGGTE------HLGLPVFNSVAEAKADTKANASVIYVPAPFAAA 129
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
AI+E +EAELDL VCITEGIPQHDM VK L Q+KTRLIGPNCPGII PGECKIGIMP
Sbjct 130 AIMEGIEAELDLIVCITEGIPQHDMVRVKHALNSQSKTRLIGPNCPGIIKPGECKIGIMP 189
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
GYIHK G IG+VSRSGTLTYEAV QTT VGLGQSTCVGIGGDPFNGT+F+DCLE+F DP
Sbjct 190 GYIHKPGKIGIVSRSGTLTYEAVFQTTAVGLGQSTCVGIGGDPFNGTNFVDCLEKFFVDP 249
Query 181 ETRG 184
+T G
Sbjct 250 QTEG 253
> At5g23250
Length=341
Score = 259 bits (662), Expect = 3e-69, Method: Compositional matrix adjust.
Identities = 129/184 (70%), Positives = 143/184 (77%), Gaps = 6/184 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
QA+ YGT++V GV P KGGT LPVF +V EAK T +AS I+VP AAA
Sbjct 71 QAIEYGTKMVAGVTPKKGGTE------HLGLPVFNTVAEAKAETKANASVIYVPAPFAAA 124
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
AI+E + AELDL VCITEGIPQHDM VK L Q+KTRLIGPNCPGII PGECKIGIMP
Sbjct 125 AIMEGLAAELDLIVCITEGIPQHDMVRVKAALNSQSKTRLIGPNCPGIIKPGECKIGIMP 184
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
GYIHK G IG+VSRSGTLTYEAV QTT VGLGQSTCVGIGGDPFNGT+F+DCLE+F DP
Sbjct 185 GYIHKPGKIGIVSRSGTLTYEAVFQTTAVGLGQSTCVGIGGDPFNGTNFVDCLEKFFVDP 244
Query 181 ETRG 184
+T G
Sbjct 245 QTEG 248
> CE05227
Length=322
Score = 249 bits (637), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 120/183 (65%), Positives = 142/183 (77%), Gaps = 6/183 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
Q L Y T++VGGVN K GT LPVFK+V EA+ TG AS I+VP + A +
Sbjct 52 QMLEYNTKVVGGVNANKAGTE------HLGLPVFKNVSEARNKTGADASVIYVPASAAGS 105
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
AI E ++AE+ L VCITEGIPQHDM VK RL +Q KTRL+GPNCPGII+ +CKIGIMP
Sbjct 106 AIEEAMDAEIPLVVCITEGIPQHDMVRVKSRLLKQNKTRLVGPNCPGIISADQCKIGIMP 165
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
G+IHK+GCIG+VSRSGTLTYEAV+QTTQVG GQ+ CVGIGGDPFNGT+FIDCL F+ DP
Sbjct 166 GHIHKRGCIGIVSRSGTLTYEAVHQTTQVGFGQTLCVGIGGDPFNGTNFIDCLNVFLEDP 225
Query 181 ETR 183
ET+
Sbjct 226 ETK 228
> CE09611
Length=321
Score = 243 bits (619), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 120/183 (65%), Positives = 140/183 (76%), Gaps = 6/183 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
Q L Y T LVGGV+P K G T LPVF SV EAK+ TG A+ I+VP A AA
Sbjct 53 QMLEYNTNLVGGVSPNKAGQT------HLGLPVFGSVAEAKDRTGADATVIYVPAAGAAR 106
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
AI E ++AE+ L V ITEGIPQ DM VK RL +Q K+RL+GPNCPGII G+CKIGIMP
Sbjct 107 AIHEAMDAEIGLIVAITEGIPQQDMVRVKNRLLKQNKSRLLGPNCPGIIASGDCKIGIMP 166
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
G+IHKKGCIG+VSRSGTLTYEAV+QTT VGLGQ+ C+GIGGDPFNGT+FIDCLE F+ D
Sbjct 167 GHIHKKGCIGIVSRSGTLTYEAVHQTTTVGLGQTRCIGIGGDPFNGTNFIDCLEVFLEDE 226
Query 181 ETR 183
+T+
Sbjct 227 QTK 229
> Hs11321581
Length=333
Score = 241 bits (614), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 132/182 (72%), Positives = 143/182 (78%), Gaps = 6/182 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
QAL YGT+LVGG P KGG T LPVF +VKEAKE TG AS I+VPP AAA
Sbjct 61 QALEYGTKLVGGTLPGKGGQTHLG------LPVFNTVKEAKEQTGATASVIYVPPPFAAA 114
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
AI E +EAE+ L VCITEGIPQ DM VK +L Q KTRLIGPNCPG+INPGECKIGIMP
Sbjct 115 AINEAIEAEIPLVVCITEGIPQQDMVRVKHKLLRQEKTRLIGPNCPGVINPGECKIGIMP 174
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
G+IHKKG IG+VSRSGTLTYEAV+QTTQVGLGQS CVGIGGDPFNGT FIDCLE F+ D
Sbjct 175 GHIHKKGRIGIVSRSGTLTYEAVHQTTQVGLGQSLCVGIGGDPFNGTDFIDCLEIFLNDS 234
Query 181 ET 182
T
Sbjct 235 AT 236
> 7292391
Length=324
Score = 236 bits (603), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 127/183 (69%), Positives = 148/183 (80%), Gaps = 6/183 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
QAL YGT+LVGG++P KGGT LPVF SV EAK+AT HA+ I+VPP AAA
Sbjct 52 QALEYGTKLVGGISPKKGGTQHLG------LPVFASVAEAKKATDPHATVIYVPPPGAAA 105
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
AI+E +EAE+ L VCITEG+PQHDM VK L Q+K+RL+GPNCPGII P +CKIGIMP
Sbjct 106 AIIEALEAEIPLIVCITEGVPQHDMVKVKHALISQSKSRLVGPNCPGIIAPEQCKIGIMP 165
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
G+IHK+G IGVVSRSGTLTYEAV+QTT+VGLGQ+ CVGIGGDPFNGT FIDCLE F+ DP
Sbjct 166 GHIHKRGKIGVVSRSGTLTYEAVHQTTEVGLGQTLCVGIGGDPFNGTDFIDCLEVFLKDP 225
Query 181 ETR 183
ET+
Sbjct 226 ETK 228
> 7300518
Length=342
Score = 223 bits (568), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 109/184 (59%), Positives = 135/184 (73%), Gaps = 6/184 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
+++ YGT +VGGVNP KGGT PVFKSV EA E A+ IF+PP AA
Sbjct 57 ESIKYGTNIVGGVNPKKGGTEHLGK------PVFKSVAEAVEKAKPDATVIFIPPPSAAE 110
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
I +E+E+ L V ITEGIPQ DM + + L Q K+RL+GPNCPGII+P +CKIGIMP
Sbjct 111 GICAAIESEIGLIVAITEGIPQADMVRISQMLNCQEKSRLLGPNCPGIISPDQCKIGIMP 170
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
G IHK+G +G+VSRSGTLTYE+V+QTT VGLGQ+ CVG+GGDPFNGTSFID L+ F++D
Sbjct 171 GDIHKRGVVGIVSRSGTLTYESVHQTTNVGLGQALCVGLGGDPFNGTSFIDALKVFLSDK 230
Query 181 ETRG 184
E +G
Sbjct 231 EIKG 234
> SPAC16E8.17c
Length=331
Score = 217 bits (553), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 113/183 (61%), Positives = 135/183 (73%), Gaps = 6/183 (3%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAA 60
A+ YGT++VGG NP K GTT PVF +++EA + T ASA+FVPP AA
Sbjct 59 HAMDYGTKVVGGTNPKKAGTTHLGK------PVFGTIEEAMKETKADASAVFVPPPLAAG 112
Query 61 AILECVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMP 120
AI E + AE+ L V ITEGIPQHDM V L+ Q+K+RL+GPNCPGII PG+CKIGIMP
Sbjct 113 AIEEAIAAEVPLIVAITEGIPQHDMLRVSDILKTQSKSRLVGPNCPGIIRPGQCKIGIMP 172
Query 121 GYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
+IHK GCIG+VSRSGTLTYEAVNQTTQ LGQS +GIGGDPF GT+FID L+ F+ DP
Sbjct 173 SHIHKPGCIGIVSRSGTLTYEAVNQTTQTDLGQSLVIGIGGDPFPGTNFIDALKLFLDDP 232
Query 181 ETR 183
T+
Sbjct 233 NTQ 235
> YOR142w
Length=329
Score = 210 bits (535), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 110/181 (60%), Positives = 129/181 (71%), Gaps = 7/181 (3%)
Query 5 YGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVKEAKEATGCHASAIFVPPAHAAAAILE 64
YGT +VGG NP K G T +GQ PVF SVK+A + TG ASAIFVPP AAAAI E
Sbjct 59 YGTNVVGGTNPKKAGQTH---LGQ---PVFASVKDAIKETGATASAIFVPPPIAAAAIKE 112
Query 65 CVEAELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPG-ECKIGIMPGYI 123
+EAE+ LAVCITEGIPQHDM + L+ Q KTRL+GPNCPGIINP + +IGI P I
Sbjct 113 SIEAEIPLAVCITEGIPQHDMLYIAEMLQTQDKTRLVGPNCPGIINPATKVRIGIQPPKI 172
Query 124 HKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADPETR 183
+ G IG++SRSGTLTYEAV QTT+ LGQS +G+GGD F GT FID L+ F+ D T
Sbjct 173 FQAGKIGIISRSGTLTYEAVQQTTKTDLGQSLVIGMGGDAFPGTDFIDALKLFLEDETTE 232
Query 184 G 184
G
Sbjct 233 G 233
> 7303012
Length=1086
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 49/165 (29%), Positives = 82/165 (49%), Gaps = 15/165 (9%)
Query 31 LPVFKSVKEA-KEATGCHASAIFVPPAHAAAAILECVE-AELDLAVCITEGIPQHDMAMV 88
+PV+K + +A + F A + LE +E ++ I EGIP++ M
Sbjct 536 IPVYKKMSDAIHKHKEVDVMVNFASMRSAYESTLEVLEFPQIRTVAIIAEGIPEN---MT 592
Query 89 KRRLREQTK--TRLIGPNCPGIINPGECKIG--------IMPGYIHKKGCIGVVSRSGTL 138
++ + E K +IGP G + PG KIG I+ +++ G + VSRSG +
Sbjct 593 RKLIIEADKKGVAIIGPATVGGVKPGCFKIGNTGGMLDNILHSKLYRPGSVAYVSRSGGM 652
Query 139 TYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADPETR 183
+ E N ++ G + IGGD + G++F+D + R+ ADPET+
Sbjct 653 SNELNNIISKATDGVIEGIAIGGDRYPGSTFMDHILRYQADPETK 697
> SPBC1703.07
Length=615
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 52/166 (31%), Positives = 79/166 (47%), Gaps = 23/166 (13%)
Query 31 LPVFKSVKEA-KEATGCHASAIFVPPAHAAAAILECVE-AELDLAVCITEGIPQHDMAMV 88
LPV+++++EA + F A A+ +E +E ++ I EG+P+
Sbjct 70 LPVYRTIEEACTKHPEVDVVVNFASSRSAYASTMELMEFPQIRCIAIIAEGVPE------ 123
Query 89 KRRLRE------QTKTRLIGPNCPGIINPGECKIG--------IMPGYIHKKGCIGVVSR 134
RR RE + +IGP G I PG KIG I+ +++ G + VS+
Sbjct 124 -RRAREILVTSKEKNVVIIGPATVGGIKPGCFKIGNTGGMMDNIVASKLYRPGSVAYVSK 182
Query 135 SGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
SG ++ E N + G + IGGD + GT+FID L RF ADP
Sbjct 183 SGGMSNELNNIISHTTDGVYEGIAIGGDRYPGTTFIDHLIRFEADP 228
> Hs22064713
Length=947
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 46/162 (28%), Positives = 80/162 (49%), Gaps = 15/162 (9%)
Query 31 LPVFKSVKEA-KEATGCHASAIFVPPAHAAAAILECVE-AELDLAVCITEGIPQHDMAMV 88
+PVFK++ +A ++ F A + +E + A++ I EGIP+ A+
Sbjct 550 IPVFKNMADAMRKHPEVDVLINFASLRSAYDSTMETMNYAQIRTIAIIAEGIPE---ALT 606
Query 89 KRRLR--EQTKTRLIGPNCPGIINPGECKIG--------IMPGYIHKKGCIGVVSRSGTL 138
++ ++ +Q +IGP G I PG KIG I+ +++ G + VSRSG +
Sbjct 607 RKLIKKADQKGVTIIGPATVGGIKPGCFKIGNTGGMLDNILASKLYRPGSVAYVSRSGGM 666
Query 139 TYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
+ E N ++ G V IGGD + G++F+D + R+ P
Sbjct 667 SNELNNIISRTTDGVYEGVAIGGDRYPGSTFMDHVLRYQDTP 708
> CE07719
Length=1099
Score = 58.2 bits (139), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 50/190 (26%), Positives = 85/190 (44%), Gaps = 19/190 (10%)
Query 9 LVGGVNPAKGGTTWTSSVGQYKL--PVFKSVKEAKEATGCHASAIFVPPAHAAAAILECV 66
+V P G GQ ++ P +KS+ +A H A + + ++ E V
Sbjct 522 VVASTYPFTGDNKQKYYFGQKEILIPAYKSMAKA---FASHPDATVMVTFASMRSVFETV 578
Query 67 -EA----ELDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIG---- 117
EA ++ + I EG+P++ + ++ E LIGP G I PG KIG
Sbjct 579 LEALQFTQIKVIAIIAEGVPENQTRKL-LKIAEDKGVTLIGPATVGGIKPGCFKIGNTGG 637
Query 118 ----IMPGYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCL 173
I+ +++ G + VSRSG ++ E N +Q G + IGGD + G+++ D +
Sbjct 638 MMDNILASKLYRPGSVAYVSRSGGMSNELNNIISQNTNGVYEGIAIGGDRYPGSTYTDHV 697
Query 174 ERFVADPETR 183
R+ D +
Sbjct 698 MRYQHDDRVK 707
> CE06997
Length=1106
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 53/191 (27%), Positives = 87/191 (45%), Gaps = 21/191 (10%)
Query 9 LVGGVNPAKGGTTWTSSVGQYKL--PVFKSVKEAKEATGCHASAI--FVPPAHAAAAILE 64
+V P G GQ ++ P +KS+ +A AT AS + F +LE
Sbjct 526 VVASTYPFTGDNKQKYYFGQKEILIPAYKSMAKAF-ATHPDASIMVTFASMRSVFETVLE 584
Query 65 CVE-AELDLAVCITEGIPQHDMAMVKRRLREQTKTR---LIGPNCPGIINPGECKIG--- 117
+E ++ + I EG+P++ R+L + R L+GP G I PG KIG
Sbjct 585 ALEFPQIKVIAIIAEGVPENQT----RKLLKIAHDRGVTLVGPATVGGIKPGCFKIGNTG 640
Query 118 -----IMPGYIHKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDC 172
I+ +++ G + VSRSG ++ E N +Q G + IGGD + G+++ D
Sbjct 641 GMMDNILASKLYRPGSVAYVSRSGGMSNELNNIISQNTNGVYEGIAIGGDRYPGSTYTDH 700
Query 173 LERFVADPETR 183
+ R+ D +
Sbjct 701 VIRYQNDDRVK 711
> Hs4501865
Length=1105
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 45/162 (27%), Positives = 79/162 (48%), Gaps = 15/162 (9%)
Query 31 LPVFKSVKEA-KEATGCHASAIFVPPAHAAAAILECVE-AELDLAVCITEGIPQHDMAMV 88
+PVFK++ +A ++ F A + +E + A++ I EGIP+ A+
Sbjct 554 IPVFKNMADAMRKHPEVDVLINFASLRSAYDSTMETMNYAQIRTIAIIAEGIPE---ALT 610
Query 89 KRRLR--EQTKTRLIGPNCPGIINPGECKIG--------IMPGYIHKKGCIGVVSRSGTL 138
++ ++ +Q +IGP G I PG KIG I+ ++ + + VSRSG +
Sbjct 611 RKLIKKADQKGVTIIGPATVGGIKPGCFKIGNTGGMLDNILASKLYPQAAVAYVSRSGGM 670
Query 139 TYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADP 180
+ E N ++ G V IGGD + G++F+D + R+ P
Sbjct 671 SNELNNIISRTTDGVYEGVAIGGDRYPGSTFMDHVLRYQDTP 712
> At5g49460
Length=608
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 59/131 (45%), Gaps = 9/131 (6%)
Query 70 LDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMPGYI------ 123
+ + I EG+P+ D + R K +IGP G I G KIG G I
Sbjct 104 IKVVAIIAEGVPESDTKQLIAYARANNKV-VIGPATVGGIQAGAFKIGDTAGTIDNIIQC 162
Query 124 --HKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADPE 181
++ G +G VS+SG ++ E N +V G + IGGD F G++ D + RF P+
Sbjct 163 KLYRPGSVGFVSKSGGMSNEMYNTVARVTDGIYEGIAIGGDVFPGSTLSDHILRFNNIPQ 222
Query 182 TRGRSTLAAHG 192
+ L G
Sbjct 223 IKMMVVLGELG 233
> At3g06650
Length=608
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 57/122 (46%), Gaps = 9/122 (7%)
Query 70 LDLAVCITEGIPQHDMAMVKRRLREQTKTRLIGPNCPGIINPGECKIGIMPGYI------ 123
+ + I EG+P+ D + R K +IGP G + G KIG G I
Sbjct 104 IKVVAIIAEGVPESDTKQLIAYARANNKV-IIGPATVGGVQAGAFKIGDTAGTIDNIIQC 162
Query 124 --HKKGCIGVVSRSGTLTYEAVNQTTQVGLGQSTCVGIGGDPFNGTSFIDCLERFVADPE 181
++ G +G VS+SG ++ E N +V G + IGGD F G++ D + RF P+
Sbjct 163 KLYRPGSVGFVSKSGGMSNEMYNTIARVTDGIYEGIAIGGDVFPGSTLSDHILRFNNIPQ 222
Query 182 TR 183
+
Sbjct 223 IK 224
> CE12804
Length=376
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 25/52 (48%), Gaps = 1/52 (1%)
Query 1 QALIYGTQLVGGVNPAKGGTTWTSSVGQYKLPVFKSVK-EAKEATGCHASAI 51
+AL+ + G+ PA GG W+S + LPV S E T C +AI
Sbjct 100 RALVSAQAFLYGLYPASGGYQWSSDIDWQPLPVHASTPGEPDLVTVCKPTAI 151
Lambda K H
0.320 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3407623970
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40