bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0268_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
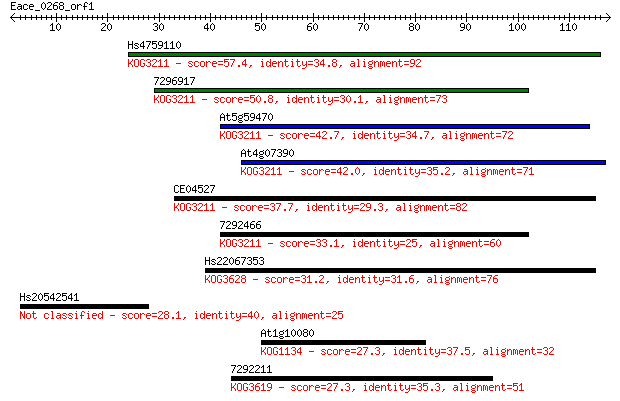
Hs4759110 57.4 6e-09
7296917 50.8 5e-07
At5g59470 42.7 2e-04
At4g07390 42.0 3e-04
CE04527 37.7 0.006
7292466 33.1 0.13
Hs22067353 31.2 0.47
Hs20542541 28.1 3.6
At1g10080 27.3 6.4
7292211 27.3 7.2
> Hs4759110
Length=247
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 55/93 (59%), Gaps = 1/93 (1%)
Query 24 YDRWEL-SSACIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTA 82
+ +W+L C+K L + I+AG+L+VK PQ+ + AKSA GLS +++ E+
Sbjct 27 FVQWDLLHVPCLKILLSKGLGLGIVAGSLLVKLPQVFKIRGAKSAEGLSLQSVMLELVAL 86
Query 83 ELVVGYNLLSKHPFSTWGEAVFIGAQNLVLLLL 115
+ Y++ + PFS+WGEA+F+ Q + + L
Sbjct 87 TGTMVYSITNNFPFSSWGEALFLMLQTITICFL 119
> 7296917
Length=197
Score = 50.8 bits (120), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 40/73 (54%), Gaps = 0/73 (0%)
Query 29 LSSACIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGY 88
L C K L + I+AG+++VK PQ+L + +KS G++ ++ ++ + Y
Sbjct 28 LDVPCFKALLSKGLGLAIIAGSVLVKVPQVLKILNSKSGEGINIVGVVLDLLAISFHLSY 87
Query 89 NLLSKHPFSTWGE 101
N + +PFS WG+
Sbjct 88 NFMHGYPFSAWGD 100
> At5g59470
Length=239
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 0/72 (0%)
Query 42 LNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGE 101
L ++A ++ VK PQ++ + KS GLS A EV + + Y L PFS +GE
Sbjct 34 LGYFLVAASMTVKLPQIMKIVDNKSVKGLSVVAFELEVIGYTISLAYCLNKDLPFSAFGE 93
Query 102 AVFIGAQNLVLL 113
F+ Q L+L+
Sbjct 94 LAFLLIQALILV 105
> At4g07390
Length=143
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 0/71 (0%)
Query 46 ILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGEAVFI 105
++A ++ VK PQ++ + KS GLS A EV + + Y L PFS +GE F+
Sbjct 38 LVAASITVKLPQIMKIVQHKSVRGLSVVAFELEVVGYTISLAYCLHKGLPFSAFGEMAFL 97
Query 106 GAQNLVLLLLY 116
Q +V L L+
Sbjct 98 LIQAVVFLNLF 108
> CE04527
Length=238
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 42/82 (51%), Gaps = 0/82 (0%)
Query 33 CIKESAGLTLNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLS 92
C K L I G++++ PQ+L + A+SA G+S + L + A Y+ S
Sbjct 28 CPKAVLSRGLGFAITLGSILLFVPQILKIQAARSAQGISAASQLLALVGAIGTASYSYRS 87
Query 93 KHPFSTWGEAVFIGAQNLVLLL 114
FS WG++ F+ Q ++++L
Sbjct 88 GFVFSGWGDSFFVAVQLVIIIL 109
> 7292466
Length=182
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 42 LNVLILAGALIVKTPQLLNVFIAKSASGLSYWAILTEVFTAELVVGYNLLSKHPFSTWGE 101
L+++ ++ L++K PQ+ + +S+ G+S + E+F+ +++ YN S + F ++ E
Sbjct 25 LSLITVSSCLVIKVPQINTIRANESSKGISVLGLCLELFSYTVMLSYNYTSGYDFLSYME 84
> Hs22067353
Length=1571
Score = 31.2 bits (69), Expect = 0.47, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 41/81 (50%), Gaps = 5/81 (6%)
Query 39 GLTLNVLILAGA-LIVKTPQLLNVFIAKSASGLSYWAIL---TEVFTA-ELVVGYNLLSK 93
G + V+ L G + KT ++ + ++ SA+G +Y+ +L VF A + G +
Sbjct 725 GANVCVVKLEGTPYLCKTDEVGEICVSSSATGTAYYGLLGITKNVFEAVPVTTGGAPIFD 784
Query 94 HPFSTWGEAVFIGAQNLVLLL 114
PF+ G FIG NLV ++
Sbjct 785 RPFTRTGLLGFIGPDNLVFIV 805
> Hs20542541
Length=164
Score = 28.1 bits (61), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 10/25 (40%), Positives = 13/25 (52%), Gaps = 0/25 (0%)
Query 3 RMRLGYVLQSCGLASAGCAAAYDRW 27
R++ Y L C L GC A +D W
Sbjct 22 RLQWAYPLWECSLLPMGCEAVFDPW 46
> At1g10080
Length=509
Score = 27.3 bits (59), Expect = 6.4, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query 50 ALIVKTPQLLNVFIAKSASGLSYWAIL--TEVFT 81
A ++ Q++NV+I K SG YW + T +F+
Sbjct 320 AYLIYKNQIINVYITKYESGGQYWPVFHNTTIFS 353
> 7292211
Length=1180
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 9/54 (16%)
Query 44 VLILAGALIVKTPQLLNVFI---AKSASGLSYWAILTEVFTAELVVGYNLLSKH 94
V ++ L++ TP++ V++ A SGL W +L F +ELV SKH
Sbjct 75 VTVIHTLLLLTTPEIKYVYVDMVAYMCSGLVIWVVLGVNFRSELV------SKH 122
Lambda K H
0.324 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40