bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0313_orf1
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
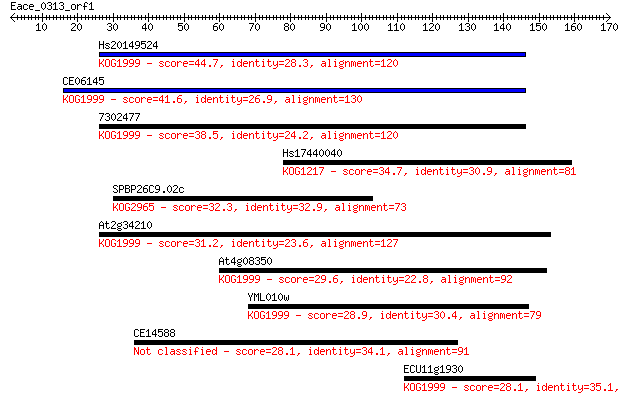
Hs20149524 44.7 8e-05
CE06145 41.6 8e-04
7302477 38.5 0.007
Hs17440040 34.7 0.084
SPBP26C9.02c 32.3 0.50
At2g34210 31.2 1.1
At4g08350 29.6 3.3
YML010w 28.9 4.5
CE14588 28.1 7.7
ECU11g1930 28.1 8.6
> Hs20149524
Length=1087
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 59/123 (47%), Gaps = 15/123 (12%)
Query 26 KKNSRPEKKFFDRDKVDSSGGQVEQG---LIPGTVKFAGLTFEEAGYIIRKIALRHLVLG 82
KK RP ++ FD +K+ S GG V LI F G + G++ + A+ V+
Sbjct 330 KKFKRPPQRLFDAEKIRSLGGDVASDGDFLI-----FEGNRYSRKGFLFKSFAMS-AVIT 383
Query 83 SAVSASLTELKEFCQNMNEADREESLTAHKPLYQMLKQQRSAFRLGDRILITQGELQGLK 142
V +L+EL++F D+ E + K++ F+ GD + + +GEL L+
Sbjct 384 EGVKPTLSELEKF------EDQPEGIDLEVVTESTGKEREHNFQPGDNVEVCEGELINLQ 437
Query 143 GRV 145
G++
Sbjct 438 GKI 440
> CE06145
Length=1208
Score = 41.6 bits (96), Expect = 8e-04, Method: Composition-based stats.
Identities = 35/131 (26%), Positives = 61/131 (46%), Gaps = 13/131 (9%)
Query 16 AARTVGGLGFKKNSRPEKKFFDRDKVDSSGGQVEQGLIPGT-VKFAGLTFEEAGYIIRKI 74
A RT +K RP K FD+D + GG++ + G + F G F G++ +
Sbjct 323 AMRTDADKNYKLKRRPMPKLFDQDTIKEVGGEI---VTDGDFLVFEGNHFRR-GFLYKYF 378
Query 75 ALRHLVLGSAVSASLTELKEFCQNMNEADREESLTAHKPLYQMLKQQRSAFRLGDRILIT 134
+ + + V +L EL++F ++ ++ RE + LK + F GD + +
Sbjct 379 PI-NAIQADGVKPTLGELEKFQESSDDLKRELETAS-------LKDTENPFVPGDIVEVK 430
Query 135 QGELQGLKGRV 145
GEL L+G+V
Sbjct 431 AGELVNLRGKV 441
> 7302477
Length=1078
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 29/120 (24%), Positives = 53/120 (44%), Gaps = 8/120 (6%)
Query 26 KKNSRPEKKFFDRDKVDSSGGQVEQGLIPGTVKFAGLTFEEAGYIIRKIALRHLVLGSAV 85
KK RP K FD + V + GG+V + F G + G++ + + +L V
Sbjct 370 KKKRRPAAKPFDPEAVRAIGGEVHSD--GDFLLFEGNRYSRKGFLYKNFTMS-AILSDGV 426
Query 86 SASLTELKEFCQNMNEADREESLTAHKPLYQMLKQQRSAFRLGDRILITQGELQGLKGRV 145
+L EL+ F ++ E + E T +F +GD + + G+L+ L+ ++
Sbjct 427 KPTLAELERFEESPEEVNLEIMGTVKDD-----PTMAHSFSMGDNVEVCVGDLENLQAKI 481
> Hs17440040
Length=542
Score = 34.7 bits (78), Expect = 0.084, Method: Composition-based stats.
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 3/84 (3%)
Query 78 HLVLGSAVSASLTELKEFCQNMNEADREESLTAHKPLYQMLKQQRSAFRLGDRILITQ-G 136
++ +G + LT LK F N +++ L+A + L Q+LK Q + F G R + ++
Sbjct 99 NIAIGVKLDEVLTPLKSFITETNCLSKQQLLSAIRQLQQLLKGQETRFAEGIRHMKSRLA 158
Query 137 ELQGLKGRV--SGLPSETKQEKDL 158
LQ GRV LP + K +
Sbjct 159 ALQNSVGRVGPDALPDGETEAKTI 182
> SPBP26C9.02c
Length=323
Score = 32.3 bits (72), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 29/73 (39%), Gaps = 9/73 (12%)
Query 30 RPEKKFFDRDKVDSSGGQVEQGLIPGTVKFAGLTFEEAGYIIRKIALRHLVLGSAVSASL 89
RP FD D D +PG GLTF EA YI +A GS V+ +
Sbjct 240 RPIHLSFDVDACDPIVAPATGTRVPG-----GLTFREAMYICESVA----ETGSLVAVDV 290
Query 90 TELKEFCQNMNEA 102
E+ N EA
Sbjct 291 MEVNPLLGNKEEA 303
> At2g34210
Length=990
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 30/131 (22%), Positives = 62/131 (47%), Gaps = 10/131 (7%)
Query 26 KKNSRPEKKFFDRDKVDSSGGQVEQGLIPGTVKF----AGLTFEEAGYIIRKIALRHLVL 81
KK P +F + D+ +VE P T + G+ F++ G++ +K++ + +
Sbjct 316 KKAFAPPPRFMNIDEARELHIRVEHRRDPMTGDYFENIGGMLFKD-GFLYKKVSTKSIA- 373
Query 82 GSAVSASLTELKEFCQNMNEADREESLTAHKPLYQMLKQQRSAFRLGDRILITQGELQGL 141
V+ + EL+ F + NE + E L+ +++ F GD +++ +G+L+ L
Sbjct 374 AQNVTPTFDELERF-KRPNE-NGEIDFVDESTLFA--NRKKGHFMKGDAVIVIKGDLKNL 429
Query 142 KGRVSGLPSET 152
KG + + E
Sbjct 430 KGWIEKVDEEN 440
> At4g08350
Length=1054
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 21/92 (22%), Positives = 49/92 (53%), Gaps = 6/92 (6%)
Query 60 AGLTFEEAGYIIRKIALRHLVLGSAVSASLTELKEFCQNMNEADREESLTAHKPLYQMLK 119
G+ F++ G+ ++++L+ + + + V+ + EL++F N + E L+
Sbjct 367 GGMLFKD-GFHYKQVSLKSITVQN-VTPTFDELEKF--NKPSENGEGDFGGLSTLFA--N 420
Query 120 QQRSAFRLGDRILITQGELQGLKGRVSGLPSE 151
+++ F GD +++ +G+L+ LKG V + E
Sbjct 421 RKKGHFMKGDAVIVIKGDLKNLKGWVEKVDEE 452
> YML010w
Length=1063
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 41/82 (50%), Gaps = 11/82 (13%)
Query 68 GYIIRKIALRHLVLGSAVSASLTELKEFCQNMNEADREESLTAHKPLYQMLKQQRSA--- 124
GY+ + ++H V + ++ EL F D LT+ + Q +K+ ++A
Sbjct 482 GYLYKSFRIQH-VETKNIQPTVEELARFGSKEGAVD----LTS---VSQSIKKAQAAKVT 533
Query 125 FRLGDRILITQGELQGLKGRVS 146
F+ GDRI + GE +G KG V+
Sbjct 534 FQPGDRIEVLNGEQRGSKGIVT 555
> CE14588
Length=490
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 31/104 (29%), Positives = 44/104 (42%), Gaps = 14/104 (13%)
Query 36 FDRDKV-DSSGGQV----EQGLIPGTVKFAGLTFEEAGYIIRKIALR--------HLVLG 82
F+ ++V D S QV L G++ + +E G I KI ++ V G
Sbjct 388 FNIERVEDRSLNQVYLRKRNTLFMGSITWQ-FIWELVGKSIEKIVIKIAGIEEYLKYVDG 446
Query 83 SAVSASLTELKEFCQNMNEADREESLTAHKPLYQMLKQQRSAFR 126
+A LT L QN + +SLT +P Y L Q S FR
Sbjct 447 GNANACLTFLAARTQNCSIVPTNKSLTIERPEYDSLSLQVSFFR 490
> ECU11g1930
Length=571
Score = 28.1 bits (61), Expect = 8.6, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 25/39 (64%), Gaps = 2/39 (5%)
Query 112 KPLYQMLKQ--QRSAFRLGDRILITQGELQGLKGRVSGL 148
+P ++ L++ Q +GD++ +T+GEL G+KG V +
Sbjct 249 EPTFEELEEFHQACPISVGDKVRVTRGELIGMKGVVRSI 287
Lambda K H
0.316 0.134 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2454498488
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40