bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
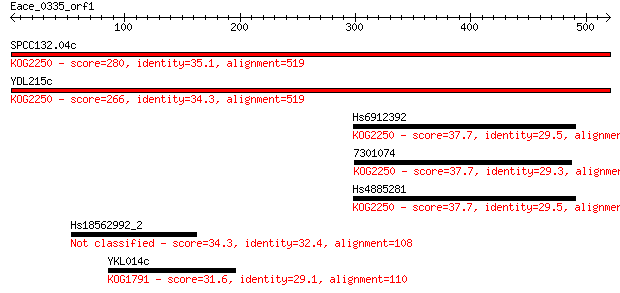
Query= Eace_0335_orf1
Length=520
Score E
Sequences producing significant alignments: (Bits) Value
SPCC132.04c 280 7e-75
YDL215c 266 8e-71
Hs6912392 37.7 0.066
7301074 37.7 0.066
Hs4885281 37.7 0.068
Hs18562992_2 34.3 0.73
YKL014c 31.6 4.6
> SPCC132.04c
Length=1106
Score = 280 bits (715), Expect = 7e-75, Method: Compositional matrix adjust.
Identities = 182/544 (33%), Positives = 270/544 (49%), Gaps = 44/544 (8%)
Query 2 RIDLAFRRGPHTEEFFSRFGDCLTMWGFYSQRKYVEPLANGSSIITTFVELLPEGILVFP 61
R+ + +RRG ++F +G +S R YVE +NG +II+ PE
Sbjct 318 RLVVGYRRGS-IFQYFPSLSKLFRYYGLHSTRTYVEQFSNGVTIIS--YNFKPELFKNAA 374
Query 62 SMPIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVGHS 121
I E + L + + + + L++ E YA F H +G
Sbjct 375 VTSINELFSQITREASLLYCLPSTDFQPLFVSEKLSIQEVTYAHCVRIFCEHVMNKLGPE 434
Query 122 FPRIEEIVRQFGGKSLTMQEIYELRTRLKTCPFSEDLIFKVVGENANIVKELYKEFASLH 181
+ + I+ ++ + + ++ RL T F+ I + + +V L+++F H
Sbjct 435 YSSLSAILDH--SNNIHAEILETIKRRLSTLAFTRTKIHDTIMQYPGLVHTLFEQFYLEH 492
Query 182 C------PRVKR---------------VLFKETNELKERILR-LESSEAVAILLKFREFT 219
P + R + L + I + + E V+++ F +F
Sbjct 493 AINHNSTPHLHRAKSATSLADEASTYSITPMSATALMDLIQKTCTNEEDVSVMEMFVKFN 552
Query 220 KSILCTNFYMPFKQGFAFRIRGSTLPSSDFPSTPCPIFFQIGGLAVGLHIRFAEVSRGGV 279
+L TNF+ K +FR S L S+ + + IG G H+RF +V+RGG+
Sbjct 553 THLLKTNFFQTTKVALSFRFDPSFLDSTQYKDPLYAMIMSIGNEFRGFHLRFRDVARGGI 612
Query 280 RLVFSVGTAAHETNRRSLLDEAYKLAFTQQFKNKDISEGGSKGIILLNKTQTLAEAKRQA 339
RL+ S A N R L DE Y LA TQ KNKDI EGG+KG+ILL K + + +
Sbjct 613 RLIKSANPEAFGLNARGLFDENYNLAKTQMLKNKDIPEGGAKGVILLGK-----DCQDKP 667
Query 340 PLAFKAYIDNMLDLLLPH--HDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSV 397
LAF YID+++DLL+ + + D LG E+ F+GPDEN T L++WA A R +
Sbjct 668 ELAFMKYIDSIIDLLIVNKSQPLVDKLGKPEILFMGPDEN--TADLVNWATIHAHRRNAP 725
Query 398 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAYVKDAGIYNKLGLKE-EEMTRIQTGGPDGD 456
WWK+F TGK GGIPHD++GMT+ SV YV+ GIY KL + + ++T++QTGGPDGD
Sbjct 726 WWKSFFTGKKPTMGGIPHDKYGMTSLSVRCYVE--GIYKKLNITDPSKLTKVQTGGPDGD 783
Query 457 LGCNALLQTKSKTIAVVDGSGVLYDPNGLDVGELHRLCSLRFEGKPTNAMLYDSSLLSPL 516
LG N + + K IAV+DGSGVLYDP GLD EL RL R +D+ LSP
Sbjct 784 LGSNEIKLSNEKYIAVIDGSGVLYDPAGLDRTELLRLADER-----KTIDHFDAGKLSPE 838
Query 517 GFKV 520
G++V
Sbjct 839 GYRV 842
> YDL215c
Length=1092
Score = 266 bits (680), Expect = 8e-71, Method: Compositional matrix adjust.
Identities = 178/544 (32%), Positives = 276/544 (50%), Gaps = 38/544 (6%)
Query 2 RIDLAFRRGPHTEEFFSRFGDCLTMWGFYSQRKYVEPLANGSSIITTFVELLPEGILVFP 61
R+ +A++R T+ ++S + + Y+E I F L E +
Sbjct 288 RLLVAYKRFT-TKRYYSALNSLFHYYKLKPSKFYLESFNVKDDDIIIFSVYLNENQQLED 346
Query 62 SM--PIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFALHFSGSVG 119
+ ++ + + L + + + + + + R + EA YA + F HF +G
Sbjct 347 VLLHDVEAALKQVEREASLLYAIPNNSFHEVYQRRQFSPKEAIYAHIGAIFINHFVNRLG 406
Query 120 HSFPRIEEIVRQFGGKSLTMQEIYELRTRLKTCPFSEDLIFKVVGENANIVKELYKEFAS 179
+ + + + ++ + L+ +L+ ++ I ++ ++ I+ +LYK FA
Sbjct 407 SDYQNLLSQITIKRNDTTLLEIVENLKRKLRNETLTQQTIINIMSKHYTIISKLYKNFAQ 466
Query 180 LHC--------------PRVKRVL-FKETNELKERILRL--ESSEAVAILLKFREFTKSI 222
+H R+++V FK E + + + S + IL F KSI
Sbjct 467 IHYYHNSTKDMEKTLSFQRLEKVEPFKNDQEFEAYLNKFIPNDSPDLLILKTLNIFNKSI 526
Query 223 LCTNFYMPFKQGFAFRIRGSTLPSS-DFPSTPCPIFFQIGGLAVGLHIRFAEVSRGGVRL 281
L TNF++ K +FR+ S + + ++P TP IFF +G G HIRF +++RGG+R+
Sbjct 527 LKTNFFITRKVAISFRLDPSLVMTKFEYPETPYGIFFVVGNTFKGFHIRFRDIARGGIRI 586
Query 282 VFSVGTAAHETNRRSLLDEAYKLAFTQQFKNKDISEGGSKGIILLNKTQTLAEAKRQAPL 341
V S ++ N ++++DE Y+LA TQQ KNKDI EGGSKG+ILLN L E Q +
Sbjct 587 VCSRNQDIYDLNSKNVIDENYQLASTQQRKNKDIPEGGSKGVILLNP--GLVEHD-QTFV 643
Query 342 AFKAYIDNMLDLLLPHHDVDDGLGI---SEVWFLGPDENTGTGGLLDWAAQRAKERGSVW 398
AF Y+D M+D+L+ ++ + + E+ F GPDE GT G +DWA A+ R W
Sbjct 644 AFSQYVDAMIDILINDPLKENYVNLLPKEEILFFGPDE--GTAGFVDWATNHARVRNCPW 701
Query 399 WKAFTTGKLIQHGGIPHDRFGMTTASVEAYVKDAGIYNKLGLKEEEMTRIQTGGPDGDLG 458
WK+F TGK GGIPHD +GMT+ V AYV IY L L + + QTGGPDGDLG
Sbjct 702 WKSFLTGKSPSLGGIPHDEYGMTSLGVRAYVN--KIYETLNLTNSTVYKFQTGGPDGDLG 759
Query 459 CNALLQTKSKT--IAVVDGSGVLYDPNGLDVGELHRLCSLRFEGKPTNAMLYDSSLLSPL 516
N +L + +A++DGSGVL DP GLD E LC L E K + +D+S LS
Sbjct 760 SNEILLSSPNECYLAILDGSGVLCDPKGLDKDE---LCRLAHERKMISD--FDTSKLSNN 814
Query 517 GFKV 520
GF V
Sbjct 815 GFFV 818
> Hs6912392
Length=558
Score = 37.7 bits (86), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 57/208 (27%), Positives = 86/208 (41%), Gaps = 32/208 (15%)
Query 298 LDEAYKLAFTQQFKNK--DISEGGSKGIILLN-KTQTLAEAKR-----QAPLAFKAYIDN 349
+DE LA +K D+ GG+K + +N K T E ++ LA K +I
Sbjct 158 VDEVKALASLMTYKCAVVDVPFGGAKAGVKINPKNYTENELEKITRRFTMELAKKGFIGP 217
Query 350 MLDLLLPHHDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQ 409
+D+ P D G E+ ++ D T G D A A TGK I
Sbjct 218 GVDVPAP----DMNTGEREMSWIA-DTYASTIGHYDINAH-----------ACVTGKPIS 261
Query 410 HGGIPHDRFGMTTASV----EAYVKDAGIYNKLGLK---EEEMTRIQTGGPDGDLGCNAL 462
GGI H R T V E ++ A + LG+ ++ +Q G G L
Sbjct 262 QGGI-HGRISATGRGVFHGIENFINQASYMSILGMTPGFRDKTFVVQGFGNVGLHSMRYL 320
Query 463 LQTKSKTIAVVDGSGVLYDPNGLDVGEL 490
+ +K IAV + G +++P+G+D EL
Sbjct 321 HRFGAKCIAVGESDGSIWNPDGIDPKEL 348
> 7301074
Length=590
Score = 37.7 bits (86), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 55/203 (27%), Positives = 83/203 (40%), Gaps = 32/203 (15%)
Query 299 DEAYKLAFTQQFKNK--DISEGGSKGIILLNKTQ----TLAEAKRQ--APLAFKAYIDNM 350
DE L+ FK D+ GG+K + +N + L + R+ LA K +I
Sbjct 152 DEVKALSALMTFKCACVDVPFGGAKAGLKINPKEYSEHELEKITRRFTLELAKKGFIGPG 211
Query 351 LDLLLPHHDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQH 410
+D+ P D G G E+ ++ D T G LD A A TGK I
Sbjct 212 VDVPAP----DMGTGEREMSWIA-DTYAKTIGHLDINAH-----------ACVTGKPINQ 255
Query 411 GGIPHDRFGMTTASV----EAYVKDAGIYNKLGLKEEEMTR---IQTGGPDGDLGCNALL 463
GGI H R T V E ++ +A +++G + +Q G G L
Sbjct 256 GGI-HGRVSATGRGVFHGLENFINEANYMSQIGTTPGWGGKTFIVQGFGNVGLHTTRYLT 314
Query 464 QTKSKTIAVVDGSGVLYDPNGLD 486
+ + I V++ G LY+P G+D
Sbjct 315 RAGATCIGVIEHDGTLYNPEGID 337
> Hs4885281
Length=558
Score = 37.7 bits (86), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 57/208 (27%), Positives = 87/208 (41%), Gaps = 32/208 (15%)
Query 298 LDEAYKLAFTQQFKNK--DISEGGSKGIILLN-KTQTLAEAKR-----QAPLAFKAYIDN 349
+DE LA +K D+ GG+K + +N K T E ++ LA K +I
Sbjct 158 VDEVKALASLMTYKCAVVDVPFGGAKAGVKINPKNYTDNELEKITRRFTMELAKKGFIGP 217
Query 350 MLDLLLPHHDVDDGLGISEVWFLGPDENTGTGGLLDWAAQRAKERGSVWWKAFTTGKLIQ 409
+D+ P D G E+ ++ D T G D A A TGK I
Sbjct 218 GIDVPAP----DMSTGEREMSWIA-DTYASTIGHYDINAH-----------ACVTGKPIS 261
Query 410 HGGIPHDRFGMTTASV----EAYVKDAGIYNKLGLKE---EEMTRIQTGGPDGDLGCNAL 462
GGI H R T V E ++ +A + LG+ ++ +Q G G L
Sbjct 262 QGGI-HGRISATGRGVFHGIENFINEASYMSILGMTPGFGDKTFVVQGFGNVGLHSMRYL 320
Query 463 LQTKSKTIAVVDGSGVLYDPNGLDVGEL 490
+ +K IAV + G +++P+G+D EL
Sbjct 321 HRFGAKCIAVGESDGSIWNPDGIDPKEL 348
> Hs18562992_2
Length=895
Score = 34.3 bits (77), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 50/121 (41%), Gaps = 25/121 (20%)
Query 54 PEGILVFPSMPIQERIDNLLSAVRLQFVMAKSKYADFARDRLLTLHEAAYAMNASHFA-- 111
P G L P P+ +++ + + K D ARDRLL AA+ ++
Sbjct 326 PSGNLSMPHKPVSDKLSEEVDEIWNDLENYIKKNEDKARDRLL----AAFPVSKDDVPDR 381
Query 112 LHFSGSVGHSFPRIEEIVRQFGG--KSLTMQEIYELRTRLKT---------CPFSEDLIF 160
LH S P E+ R G +L++ E L T +K+ CPF EDLI
Sbjct 382 LH-----AESTP---ELSRDVGRSVSTLSLPESQALLTPVKSRAGRASRANCPFEEDLIS 433
Query 161 K 161
K
Sbjct 434 K 434
> YKL014c
Length=1764
Score = 31.6 bits (70), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 32/131 (24%), Positives = 56/131 (42%), Gaps = 21/131 (16%)
Query 86 KYADFARDRLLTLHEAAYAMNASHFALHFSGSV----GHSFPRIEEIVRQ--FGGKSL-- 137
+Y +FA +L A M S F + G+V GH EI+++ F G+++
Sbjct 844 RYINFALPKLAIEKRNALLMKKSRFMCNLIGAVCFETGHQLVEFREIIQKVVFSGENVEE 903
Query 138 --TMQEIYE----------LRTRLKTCPFSEDLIFKVVGENA-NIVKELYKEFASLHCPR 184
E+Y+ + L T + L+ E+ NI+++L+ E ++
Sbjct 904 YANYNELYQKEDVNAFLTSVSEYLSTSALTSLLMCSTKLESTRNILQKLFNEGKTIKISL 963
Query 185 VKRVLFKETNE 195
VK +L K NE
Sbjct 964 VKNILNKAANE 974
Lambda K H
0.322 0.139 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14139394082
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40