bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0346_orf2
Length=253
Score E
Sequences producing significant alignments: (Bits) Value
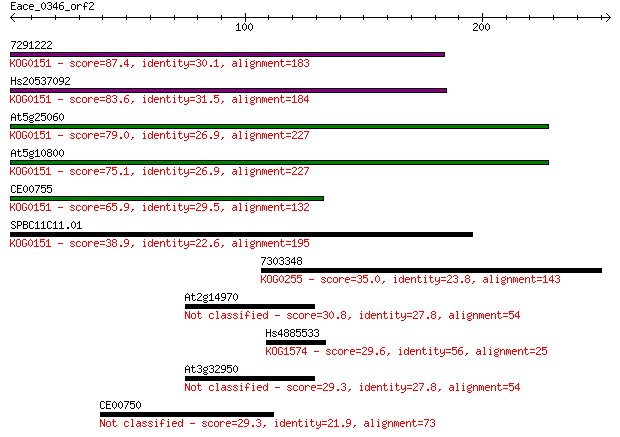
7291222 87.4 3e-17
Hs20537092 83.6 3e-16
At5g25060 79.0 8e-15
At5g10800 75.1 1e-13
CE00755 65.9 7e-11
SPBC11C11.01 38.9 0.011
7303348 35.0 0.15
At2g14970 30.8 3.1
Hs4885533 29.6 6.3
At3g32950 29.3 7.9
CE00750 29.3 8.9
> 7291222
Length=957
Score = 87.4 bits (215), Expect = 3e-17, Method: Composition-based stats.
Identities = 55/183 (30%), Positives = 92/183 (50%), Gaps = 18/183 (9%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
L+R++T +RA I AM FC+ H ++ E+C C+ ESL+ +T + ++AR+YL+SD+L N
Sbjct 488 LIRHLTPERARIGDAMIFCIEHADAADEICECIAESLSNINTLASKKIARLYLISDILHN 547
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
+ + A+A +R +EK L I D L + L + R + E G + +
Sbjct 548 CTVKVANASFFRKSVEKQLLDIFDN----LHNYYLNI-----ESRLKAE---GFKSRVCN 595
Query 121 MTKLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLDGE 180
+ + WE W +YP F+ + A G+ + + A E E D +DG
Sbjct 596 VIRTWEEWTIYPKDFMAQLTAKFLGKPYVKPVNNSPQAE------ETRFDEALDEDIDGA 649
Query 181 PLA 183
PL+
Sbjct 650 PLS 652
> Hs20537092
Length=1029
Score = 83.6 bits (205), Expect = 3e-16, Method: Composition-based stats.
Identities = 58/186 (31%), Positives = 91/186 (48%), Gaps = 22/186 (11%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T + I AM FC+++ ++ E+ C+TESL++ T L ++AR+YL+SD+L N
Sbjct 545 ILRGLTPRKNDIGDAMVFCLNNAEAAEEIVDCITESLSILKTPLPKKIARLYLVSDVLYN 604
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
SSA+ A+A YR E L ++ DL A+ RT Q Q
Sbjct 605 SSAKVANASYYRKFFETKL-------CQIFSDL-------NATYRTIQGHLQSENFKQRV 650
Query 121 MT--KLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLD 178
MT + WE W +YP FL ++ G I++ + + A +E +LD
Sbjct 651 MTCFRAWEDWAIYPEPFLIKLQNIFLG---LVNIIEEKETEDVPDDLDGAPIE---EELD 704
Query 179 GEPLAE 184
G PL +
Sbjct 705 GAPLED 710
> At5g25060
Length=944
Score = 79.0 bits (193), Expect = 8e-15, Method: Composition-based stats.
Identities = 61/232 (26%), Positives = 113/232 (48%), Gaps = 30/232 (12%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T +R+ I AM F + + ++ EV LTESLTL +T++ ++AR+ L+SD+L N
Sbjct 446 MLRALTLERSQIKEAMGFALDNADAAGEVVEVLTESLTLKETSIPTKVARLMLVSDILHN 505
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYK 120
SSA+ +A YR E LP I + ++ + + + R E + ER L K
Sbjct 506 SSARVKNASAYRTKFEATLPDIMESFNDLYRSI---------TGRITAEALK--ERVL-K 553
Query 121 MTKLWEAWGVYPPAFLRGIEATLCGRDITATILDPEAAAAAAAKCEAAALELADPQLDGE 180
+ ++W W ++ A++ G+ +T ++ + + C A P+++ +
Sbjct 554 VLQVWADWFLFSDAYIYGLRSTFLRSGVSGV-------TSFHSICGDA------PEIENK 600
Query 181 PLAENLRPL-----IAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGLGVLPTR 227
A+N+ + A + A E + L +L+R C+ GL ++ R
Sbjct 601 SYADNMSDIGKINPDAALAIGKGAARQELMNLPIAELERRCRHNGLSLVGGR 652
> At5g10800
Length=957
Score = 75.1 bits (183), Expect = 1e-13, Method: Composition-based stats.
Identities = 61/242 (25%), Positives = 111/242 (45%), Gaps = 24/242 (9%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
+LR +T +R+ I AM F + + ++ EV LTESLTL +T++ ++AR+ L+SD++ N
Sbjct 439 MLRALTLERSQIREAMGFALDNAEAAGEVVEVLTESLTLKETSIPTKVARLMLVSDIIHN 498
Query 61 SSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDL--RLGVGVGGASPRTQQEQW------- 111
SSA+ +A YR E LP I + ++ + + R+ P ++ +
Sbjct 499 SSARVKNASAYRTKFEATLPDIMESFNDLYHSVHGRITAEALRVIPESKYSSYVVLYPLK 558
Query 112 -----QGVERALYKMTKLWEAWGVYPPAFLRGIEAT-LCGRDITATILDPEAAAAAAAKC 165
V+ + K+ ++W W ++ A++ G+ AT L R+ T A +
Sbjct 559 FCSHSYFVQERVLKVLQVWADWFLFSDAYINGLRATFLRSRNFGVTSFHSICGDAPDIEK 618
Query 166 EAAALELADPQLDGEPLAENLRPLIAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGLGVLP 225
+ + D + A + A+ L RP + +L+R C+ GL +L
Sbjct 619 KGLIGNMNDADKINQDAALAMGEGAARQELMNRPIS---------ELERRCRHNGLSLLG 669
Query 226 TR 227
R
Sbjct 670 GR 671
> CE00755
Length=933
Score = 65.9 bits (159), Expect = 7e-11, Method: Composition-based stats.
Identities = 39/135 (28%), Positives = 71/135 (52%), Gaps = 17/135 (12%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCLTESLTLPDTALTMRLARMYLLSDLLAN 60
L+R +T ++ SI A M +C+ + +AE+C C+ +SLT+ D L +++R+YL++D+L+N
Sbjct 533 LIRELTPEKTSIAATMVWCIQNAKYAAEICECVYDSLTIEDIPLYKKISRLYLINDILSN 592
Query 61 SSAQTAH-AWTYRHHLEKHLPFIADRWSKVLEDLRLGVGVG--GASPRTQQEQWQGVERA 117
+ + YR H E +LE + + VG R +QEQ++
Sbjct 593 CVQRNVRDVFLYRSHFE-----------ALLEKIFVAVGKAYRAIPSRIKQEQFKQRVMC 641
Query 118 LYKMTKLWEAWGVYP 132
+++ +E VYP
Sbjct 642 VFRN---FEEMAVYP 653
> SPBC11C11.01
Length=493
Score = 38.9 bits (89), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 44/218 (20%), Positives = 87/218 (39%), Gaps = 36/218 (16%)
Query 1 LLRNITRDRASICAAMSFCMSHGSSSAEVCSCL----------TESLTLPDTALTMR--- 47
L++ + ++ SI + F + H ++ E+ ++ D + R
Sbjct 247 LIQGLNCEKGSIGNLLCFAVEHVNNHVEITDAFLKEFFNDQPTDDTKVYDDDYINERGEK 306
Query 48 -LARMYLLSDLLANSSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRL-GVGVGGASPR 105
L+ +YL++D+L N + T+ W YR E H+ ++L+DL L +GG R
Sbjct 307 KLSLIYLMNDILFNGISGTSLVWRYRFSFEPHV-------ERLLDDLYLFSKRLGG---R 356
Query 106 TQQEQWQGVERALYKMTKLWEAWGVYPPAFLRGIEATLCGR-----DITATILDPEAAAA 160
+++ + + + K+ ++W+ W + L G I + L E +
Sbjct 357 IKEDIFC---KKVVKVIEVWKTWIAFQEETLERAWRNFSGNTPQSPQINSAALKVETKNS 413
Query 161 AAAKCEAAALELADPQLDGEPLAEN---LRPLIAQFPL 195
A E D + +G P+ N I+Q P+
Sbjct 414 WTAISEETEGLQDDEEYNGIPVDVNELLNVEFISQIPM 451
> 7303348
Length=561
Score = 35.0 bits (79), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 34/146 (23%), Positives = 61/146 (41%), Gaps = 16/146 (10%)
Query 107 QQEQWQGVERALYKMTKLWEAWGVYPPAFLRGIEATLCG---RDITATILDPEAAAAAAA 163
Q W + + + GVYP AF+ G+E + G R++++ +L+ A A
Sbjct 207 QALSWDYISFLFFALLNAVGTSGVYPLAFIIGVE--MVGPRKREMSSIVLNYFYAVGEAL 264
Query 164 KCEAAALELADPQLDGEPLAENLRPLIAQFPLWLRPAAMEWLLLDRYQLDRLCQQRGLGV 223
L + P LA ++ PLI WL P ++ WLL ++ GV
Sbjct 265 ----LGLSVFLPDWRQLQLALSVPPLICVAYFWLVPESVRWLLARN-------RREQAGV 313
Query 224 LPTRSSSSSSSERRQKQLVQLARQEL 249
+ R++ + + + + +QEL
Sbjct 314 IIRRAAKVNRRDISVELMASFKQQEL 339
> At2g14970
Length=771
Score = 30.8 bits (68), Expect = 3.1, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 75 LEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYKMTKLWEAW 128
+ +H+P +RW+K+ E++R + + E++Q V L +M LW +W
Sbjct 227 VREHVPITINRWTKIGEEIRTLLWKSVQAKFELDEEYQKVA-VLKQMGCLWRSW 279
> Hs4885533
Length=435
Score = 29.6 bits (65), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 3/28 (10%)
Query 109 EQWQGVERALYKMT---KLWEAWGVYPP 133
E+W+G ER L +T KLW+AWG P
Sbjct 81 EKWRGSERVLPPLTRILKLWKAWGDEQP 108
> At3g32950
Length=787
Score = 29.3 bits (64), Expect = 7.9, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Query 75 LEKHLPFIADRWSKVLEDLRLGVGVGGASPRTQQEQWQGVERALYKMTKLWEAW 128
+ +H+P D W+K+ E++R + + E++Q V L +M LW +W
Sbjct 124 VREHVPITIDHWTKIGEEIRTLLWKSIQARFELDEEYQKVA-VLKQMGCLWRSW 176
> CE00750
Length=419
Score = 29.3 bits (64), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/73 (21%), Positives = 34/73 (46%), Gaps = 0/73 (0%)
Query 39 LPDTALTMRLARMYLLSDLLANSSAQTAHAWTYRHHLEKHLPFIADRWSKVLEDLRLGVG 98
P+ + +MY +++L+A + +T TY+ ++EK L E L++ G
Sbjct 208 FPNAEAVKKFGQMYSMTELIAETWLETRRQQTYQKYVEKGLQTDPSVIQFYCESLKIAEG 267
Query 99 VGGASPRTQQEQW 111
+ +P Q + +
Sbjct 268 LRLENPSIQTKSF 280
Lambda K H
0.321 0.132 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5223052002
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40