bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0374_orf1
Length=263
Score E
Sequences producing significant alignments: (Bits) Value
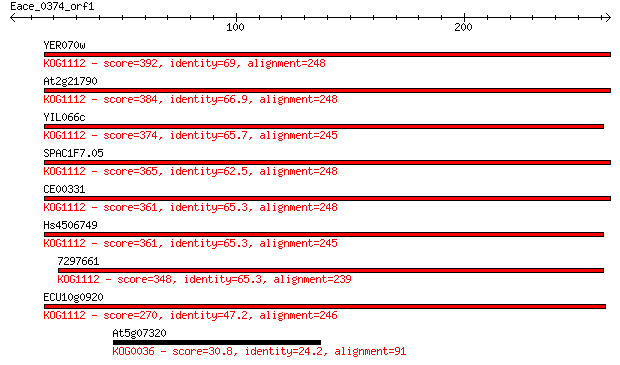
YER070w 392 3e-109
At2g21790 384 1e-106
YIL066c 374 9e-104
SPAC1F7.05 365 6e-101
CE00331 361 7e-100
Hs4506749 361 7e-100
7297661 348 8e-96
ECU10g0920 270 2e-72
At5g07320 30.8 2.8
> YER070w
Length=888
Score = 392 bits (1008), Expect = 3e-109, Method: Compositional matrix adjust.
Identities = 171/248 (68%), Positives = 209/248 (84%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ DVY V++ KD++N A+VYDRDF Y YF FKTLERSYLLR G + ERPQH +MRV
Sbjct 115 MISDDVYNIVMENKDKLNSAIVYDRDFQYSYFGFKTLERSYLLRINGQVAERPQHLIMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A GIH D++ LETY LMS K+F HA+PTLFNAGTP+PQMSSCFL+ MKEDSI+GI+ T
Sbjct 175 ALGIHGRDIEAALETYNLMSLKYFTHASPTLFNAGTPKPQMSSCFLVAMKEDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L++CALISKTAGG+GL + +IR+TGSYI GTNG SNGLIPM+RVFN+ +RYVDQGG KR
Sbjct 235 LKECALISKTAGGIGLHIHNIRSTGSYIAGTNGTSNGLIPMIRVFNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWHADIF+F++IRKNHGKE++RARDLFPALW+PDLFM+RV ENG+WTL P
Sbjct 295 GAFALYLEPWHADIFDFIDIRKNHGKEEIRARDLFPALWIPDLFMKRVEENGTWTLFSPT 354
Query 256 ECPGLTSC 263
PGL+ C
Sbjct 355 SAPGLSDC 362
> At2g21790
Length=816
Score = 384 bits (986), Expect = 1e-106, Method: Compositional matrix adjust.
Identities = 166/248 (66%), Positives = 208/248 (83%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
L+ DV+ +++ ++ ++YDRDF+YDYF FKTLERSYLL+ G +VERPQH LMRV
Sbjct 115 LIADDVFEIIMQNAARLDSEIIYDRDFEYDYFGFKTLERSYLLKVQGTVVERPQHMLMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A GIH D+ ++TY LMS ++F HA+PTLFNAGTPRPQ+SSCFL+ MK+DSI+GI+ T
Sbjct 175 AVGIHKDDIDSVIQTYHLMSQRWFTHASPTLFNAGTPRPQLSSCFLVCMKDDSIEGIYET 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L++CA+ISK+AGG+G+SV +IRATGSYIRGTNG SNG++PMLRVFND +RYVDQGGGKRK
Sbjct 235 LKECAVISKSAGGIGVSVHNIRATGSYIRGTNGTSNGIVPMLRVFNDTARYVDQGGGKRK 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ AVY+EPWHAD++EFLE+RKNHGKE+ RARDLF ALW+PDLFM+RV NG W+L CP
Sbjct 295 GAFAVYLEPWHADVYEFLELRKNHGKEEHRARDLFYALWLPDLFMERVQNNGQWSLFCPN 354
Query 256 ECPGLTSC 263
E PGL C
Sbjct 355 EAPGLADC 362
> YIL066c
Length=869
Score = 374 bits (961), Expect = 9e-104, Method: Compositional matrix adjust.
Identities = 161/245 (65%), Positives = 203/245 (82%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ ++Y V++ KD +N A+VYDRDF Y YF FKTLERSYLLR G + ERPQH +MRV
Sbjct 115 MISDEIYNIVMENKDTLNSAIVYDRDFQYTYFGFKTLERSYLLRLNGEVAERPQHLVMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A GIH D++ L+TY LMS ++F HA+PTLFNAGTP PQMSSCFL+ MK+DSI+GI+ T
Sbjct 175 ALGIHGSDIESVLKTYNLMSLRYFTHASPTLFNAGTPHPQMSSCFLIAMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L++CA+ISKTAGG+GL + +IR+TGSYI GTNG SNGLIPM+RVFN+ +RYVDQGG KR
Sbjct 235 LKECAMISKTAGGVGLHINNIRSTGSYIAGTNGTSNGLIPMIRVFNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+++EPWHADIF+F++IRK HGKE++RARDLFPALW+PDLFM+RV E+G WTL P
Sbjct 295 GAFALFLEPWHADIFDFVDIRKTHGKEEIRARDLFPALWIPDLFMKRVQEDGPWTLFSPS 354
Query 256 ECPGL 260
PGL
Sbjct 355 AAPGL 359
> SPAC1F7.05
Length=811
Score = 365 bits (936), Expect = 6e-101, Method: Compositional matrix adjust.
Identities = 155/248 (62%), Positives = 200/248 (80%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ +Y V+K+KDE++ A++YDRDF Y++F FKTLERSYLLR G + ERPQH +MRV
Sbjct 115 MISDKIYDIVMKHKDELDSAIIYDRDFTYNFFGFKTLERSYLLRIDGKVAERPQHMIMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A GIH D++ +ETY LMS ++F HA+PTLFNAGTPRPQ+SSCFL+TMK+DSI+GI+ T
Sbjct 175 AVGIHGEDIEAAIETYNLMSQRYFTHASPTLFNAGTPRPQLSSCFLVTMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+ CA+ISKTAGG+G+++ +IRATGSYI GTNG SNG++PM+RV+N+ +RYVDQGG KR
Sbjct 235 LKMCAMISKTAGGIGINIHNIRATGSYIAGTNGTSNGIVPMIRVYNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A Y+EPWHAD+ +FLE+RK HG E RAR++F ALW+PDLFMQRV N WT CP
Sbjct 295 GAFAAYLEPWHADVMDFLELRKTHGNEDFRAREMFYALWIPDLFMQRVERNEQWTFFCPN 354
Query 256 ECPGLTSC 263
E PGL
Sbjct 355 EAPGLADV 362
> CE00331
Length=788
Score = 361 bits (927), Expect = 7e-100, Method: Compositional matrix adjust.
Identities = 162/248 (65%), Positives = 198/248 (79%), Gaps = 0/248 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
++ + +A + K D++N A+VYDRD+ Y YF FKTLERSYLL+ IVERPQ LMRV
Sbjct 121 MISDETWAIIEKNADKLNSAIVYDRDYSYTYFGFKTLERSYLLKINKEIVERPQQMLMRV 180
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ +ETY LMS ++ HA+PTLFN+GT RPQMSSCFLLTM EDSI GI+ T
Sbjct 181 SIGIHGDDITSAIETYNLMSERYMTHASPTLFNSGTCRPQMSSCFLLTMSEDSILGIYDT 240
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISK+AGG+GL+V IRATGS I GTNG SNGLIPMLRV+N+ +RYVDQGG KR
Sbjct 241 LKQCALISKSAGGIGLNVHKIRATGSVIAGTNGTSNGLIPMLRVYNNTARYVDQGGNKRP 300
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWHADIFEF+ +RKN G E+ RARDLF ALW+PDLFM+RV ++ W+LMCPC
Sbjct 301 GAFAIYLEPWHADIFEFVSLRKNTGPEEERARDLFLALWIPDLFMKRVEKDQEWSLMCPC 360
Query 256 ECPGLTSC 263
ECPGL C
Sbjct 361 ECPGLDDC 368
> Hs4506749
Length=792
Score = 361 bits (927), Expect = 7e-100, Method: Compositional matrix adjust.
Identities = 160/245 (65%), Positives = 200/245 (81%), Gaps = 0/245 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
+V V+ KD +N A++YDRDF Y+YF FKTLERSYLL+ G + ERPQH LMRV
Sbjct 115 MVAKSTLDIVLANKDRLNSAIIYDRDFSYNYFGFKTLERSYLLKINGKVAERPQHMLMRV 174
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
+ GIH D+ +ETY L+S ++F HA+PTLFNAGT RPQ+SSCFLL+MK+DSI+GI+ T
Sbjct 175 SVGIHKEDIDAAIETYNLLSERWFTHASPTLFNAGTNRPQLSSCFLLSMKDDSIEGIYDT 234
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
L+QCALISK+AGG+G++V+ IRATGSYI GTNG SNGL+PMLRV+N+ +RYVDQGG KR
Sbjct 235 LKQCALISKSAGGIGVAVSCIRATGSYIAGTNGNSNGLVPMLRVYNNTARYVDQGGNKRP 294
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
G+ A+Y+EPWH DIFEFL+++KN GKE+ RARDLF ALW+PDLFM+RV N W+LMCP
Sbjct 295 GAFAIYLEPWHLDIFEFLDLKKNTGKEEQRARDLFFALWIPDLFMKRVETNQDWSLMCPN 354
Query 256 ECPGL 260
ECPGL
Sbjct 355 ECPGL 359
> 7297661
Length=771
Score = 348 bits (892), Expect = 8e-96, Method: Compositional matrix adjust.
Identities = 156/239 (65%), Positives = 192/239 (80%), Gaps = 0/239 (0%)
Query 22 YAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRVACGIHC 81
Y V K +N +++YDRDF Y+YF FKTLERSYLL+ G I ERPQH LMRVA GIH
Sbjct 91 YNVVKKNATRLNSSIIYDRDFGYNYFGFKTLERSYLLKRNGKIAERPQHMLMRVAIGIHG 150
Query 82 GDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFSTLRQCAL 141
D+ +ETY L+S ++F HA+PTLF A T RPQ+SSCFLLTM DSI+GIF ++ QCA+
Sbjct 151 EDIDAAVETYNLLSERYFTHASPTLFAAATNRPQLSSCFLLTMTADSIEGIFKSVEQCAM 210
Query 142 ISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRKGSLAVY 201
ISK+AGG+GL+V IRA G+ I GTNG SNGL+PMLRVFN+ +RYVDQGGGKR G+ A+Y
Sbjct 211 ISKSAGGIGLNVHCIRAKGTSICGTNGTSNGLVPMLRVFNNVARYVDQGGGKRPGAFAIY 270
Query 202 VEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPCECPGL 260
+EPWH+D+FEFLE++KN GKE+ RARDLF ALW+PDLFM+RV NG W+LMCP +CPGL
Sbjct 271 LEPWHSDVFEFLELKKNTGKEENRARDLFYALWIPDLFMKRVEANGDWSLMCPHKCPGL 329
> ECU10g0920
Length=768
Score = 270 bits (690), Expect = 2e-72, Method: Compositional matrix adjust.
Identities = 116/246 (47%), Positives = 173/246 (70%), Gaps = 0/246 (0%)
Query 16 LVHADVYAFVVKYKDEINKALVYDRDFDYDYFAFKTLERSYLLRAGGVIVERPQHTLMRV 75
+V ++Y ++++ + I + Y+ DF + YF TL +SYL++ G I+ERPQ MRV
Sbjct 109 MVSEEMYGIILEHGETIEGMINYENDFLFSYFGILTLMKSYLIKVGEEIIERPQDMFMRV 168
Query 76 ACGIHCGDLQKTLETYELMSCKFFIHATPTLFNAGTPRPQMSSCFLLTMKEDSIDGIFST 135
A IH D +K E Y L+S +F HATPTL+N+ PQ++SCFL+T +EDSI+G++
Sbjct 169 ALQIHKTDFEKVREVYNLLSGHYFTHATPTLYNSCLKNPQLASCFLITPREDSIEGVYHM 228
Query 136 LRQCALISKTAGGLGLSVTDIRATGSYIRGTNGYSNGLIPMLRVFNDASRYVDQGGGKRK 195
+ Q A+I+K +GG+GL++ IR+ GS +R T G SNG+IP+++V N RY++QG +R
Sbjct 229 INQAAIITKYSGGIGLNLHGIRSKGSSLRSTGGRSNGIIPLIQVLNATKRYINQGAERRP 288
Query 196 GSLAVYVEPWHADIFEFLEIRKNHGKEKMRARDLFPALWVPDLFMQRVYENGSWTLMCPC 255
GS+A+++EPWH +IF+FLE+RKN G E+ RARD+F ALW+ DLFM+RV N W+L CP
Sbjct 289 GSIAIFLEPWHMEIFDFLELRKNTGPEEFRARDIFTALWINDLFMERVKNNEEWSLFCPS 348
Query 256 ECPGLT 261
+ GL+
Sbjct 349 QAVGLS 354
> At5g07320
Length=479
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 22/91 (24%), Positives = 39/91 (42%), Gaps = 14/91 (15%)
Query 46 YFAFKTLERSYLLRAGGVIVERPQHTLMRVACGIHCGDLQKTLETYELMSCKFFIHATPT 105
Y K L R+Y+L+ + L++++CG+ G L SC + + T
Sbjct 375 YETLKDLSRTYILQ------DTEPGPLIQLSCGMTSGALGA--------SCVYPLQVVRT 420
Query 106 LFNAGTPRPQMSSCFLLTMKEDSIDGIFSTL 136
A + + M F+ TMK + + G + L
Sbjct 421 RMQADSSKTTMKQEFMNTMKGEGLRGFYRGL 451
Lambda K H
0.326 0.141 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5578361662
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40