bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0387_orf3
Length=203
Score E
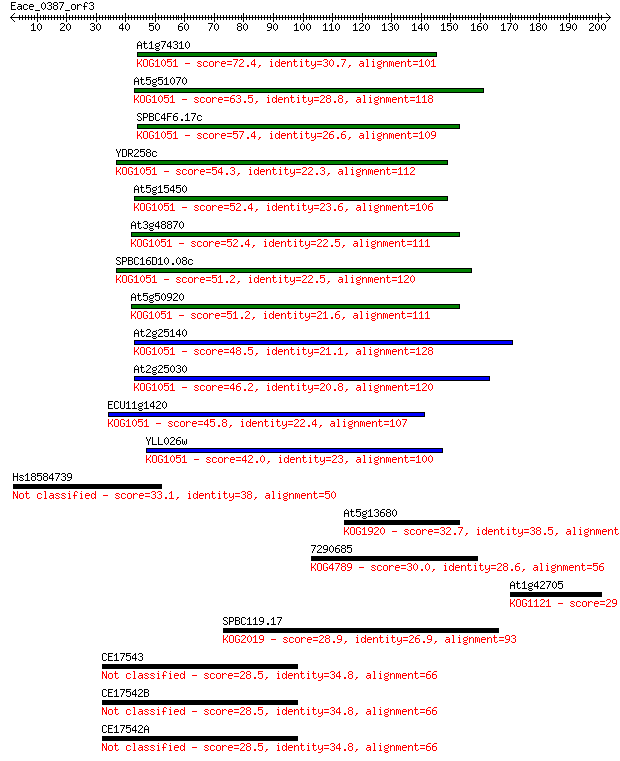
Sequences producing significant alignments: (Bits) Value
At1g74310 72.4 5e-13
At5g51070 63.5 2e-10
SPBC4F6.17c 57.4 2e-08
YDR258c 54.3 1e-07
At5g15450 52.4 6e-07
At3g48870 52.4 7e-07
SPBC16D10.08c 51.2 1e-06
At5g50920 51.2 1e-06
At2g25140 48.5 1e-05
At2g25030 46.2 5e-05
ECU11g1420 45.8 6e-05
YLL026w 42.0 7e-04
Hs18584739 33.1 0.39
At5g13680 32.7 0.53
7290685 30.0 3.3
At1g42705 29.6 4.3
SPBC119.17 28.9 7.4
CE17543 28.5 9.4
CE17542B 28.5 9.6
CE17542A 28.5 9.6
> At1g74310
Length=911
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 63/101 (62%), Gaps = 0/101 (0%)
Query 44 RQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKVTD 103
R VM+EV +F+P+++ R+ EI++F+PL+ L+ V L++ + LA +GV VTD
Sbjct 740 RDCVMREVRKHFRPELLNRLDEIVVFDPLSHDQLRKVARLQMKDVAVRLAERGVALAVTD 799
Query 104 SAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKV 144
+A+ +IL + YG R + +++EK + ++ +++ ++
Sbjct 800 AALDYILAESYDPVYGARPIRRWMEKKVVTELSKMVVREEI 840
> At5g51070
Length=945
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 65/119 (54%), Gaps = 1/119 (0%)
Query 43 MRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKVT 102
M+ V++E+ YF+P+++ R+ EI+IF L A + +L+L L L L A GV +V+
Sbjct 808 MKALVVEELKNYFRPELLNRIDEIVIFRQLEKAQMMEILNLMLQDLKSRLVALGVGLEVS 867
Query 103 DSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVLARSNT-NPN 160
+ I ++ YG R + + + + ++ ++ ++G K GD A + +T NP+
Sbjct 868 EPVKELICKQGYDPAYGARPLRRTVTEIVEDPLSEAFLAGSFKPGDTAFVVLDDTGNPS 926
> SPBC4F6.17c
Length=803
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 62/109 (56%), Gaps = 0/109 (0%)
Query 44 RQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKVTD 103
R +VM V Y+ P+ + R+ + I+F L+ L+ +++++L + + L + + VT+
Sbjct 672 RDAVMDVVQKYYPPEFLNRIDDQIVFNKLSEKNLEDIVNVRLDEVQQRLNDRRIILTVTE 731
Query 104 SAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVL 152
+A ++ E+ S YG R + + ++K I +A +I G++KS + V+
Sbjct 732 AARKWLAEKGYSPAYGARPLNRLIQKRILNTMAMKIIQGEIKSDENVVI 780
> YDR258c
Length=811
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/112 (22%), Positives = 60/112 (53%), Gaps = 0/112 (0%)
Query 37 GDILKRMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKG 96
G I + V++ + + P+ I R+ +I++F L+ L+ ++ +++A + LA K
Sbjct 671 GKIDTATKNKVIEAMKRSYPPEFINRIDDILVFNRLSKKVLRSIVDIRIAEIQDRLAEKR 730
Query 97 VDFKVTDSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGD 148
+ +TD A ++ ++ YG R + + + + I +A L+ G++++G+
Sbjct 731 MKIDLTDEAKDWLTDKGYDQLYGARPLNRLIHRQILNSMATFLLKGQIRNGE 782
> At5g15450
Length=968
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 57/106 (53%), Gaps = 0/106 (0%)
Query 43 MRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKVT 102
+++ VM F+P+ + R+ E I+F+PL+ + ++ L+LA + + +A + + +T
Sbjct 821 IKERVMNAARSIFRPEFMNRVDEYIVFKPLDREQINRIVRLQLARVQKRIADRKMKINIT 880
Query 103 DSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGD 148
D+A+ + YG R + + +++ I+ +A ++ G K D
Sbjct 881 DAAVDLLGSLGYDPNYGARPVKRVIQQNIENELAKGILRGDFKEED 926
> At3g48870
Length=952
Score = 52.4 bits (124), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 25/111 (22%), Positives = 58/111 (52%), Gaps = 0/111 (0%)
Query 42 RMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKV 101
R++ V +E+ YF+P+ + R+ E+I+F L +K + + L + L K ++ +V
Sbjct 808 RIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADIMLKEVVARLEVKEIELQV 867
Query 102 TDSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVL 152
T+ +++ YG R + + + + ++ +A ++S +K GD ++
Sbjct 868 TERFKERVVDEGFDPSYGARPLRRAIMRLLEDSMAEKMLSRDIKEGDSVIV 918
> SPBC16D10.08c
Length=905
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/122 (22%), Positives = 63/122 (51%), Gaps = 2/122 (1%)
Query 37 GDILKRMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAA-- 94
G I R+ VM + G+F+P+ + R+S I+IF L ++ ++ ++ + + L +
Sbjct 750 GKIDSTTREMVMNSIRGFFRPEFLNRISSIVIFNRLRRVDIRNIVENRILEVQKRLQSNH 809
Query 95 KGVDFKVTDSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVLAR 154
+ + +V+D A + S YG R + + ++ + +A L+++G+++ + A +
Sbjct 810 RSIKIEVSDEAKDLLGSAGYSPAYGARPLNRVIQNQVLNPMAVLILNGQLRDKETAHVVV 869
Query 155 SN 156
N
Sbjct 870 QN 871
> At5g50920
Length=929
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/111 (21%), Positives = 60/111 (54%), Gaps = 0/111 (0%)
Query 42 RMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKV 101
R++ V +E+ YF+P+ + R+ E+I+F L +K + + L + L K ++ +V
Sbjct 787 RIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKLEVKEIADILLKEVFERLKKKEIELQV 846
Query 102 TDSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVL 152
T+ +++ + YG R + + + + ++ +A +++ ++K GD ++
Sbjct 847 TERFKERVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIV 897
> At2g25140
Length=874
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/128 (21%), Positives = 65/128 (50%), Gaps = 1/128 (0%)
Query 43 MRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKVT 102
M++ V++ F+P+ + R+ E I+F+PL++ + ++ L++ + SL K + + T
Sbjct 738 MKRQVVELARQNFRPEFMNRIDEYIVFQPLDSNEISKIVELQMRRVKNSLEQKKIKLQYT 797
Query 103 DSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVLARSNTNPNQL 162
A+ + + YG R + + +++ ++ +A ++ G D VL + +
Sbjct 798 KEAVDLLAQLGFDPNYGARPVKRVIQQMVENEIAVGILKGDFAEED-TVLVDVDHLASDN 856
Query 163 NLIICELD 170
L+I +L+
Sbjct 857 KLVIKKLE 864
> At2g25030
Length=265
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/120 (20%), Positives = 61/120 (50%), Gaps = 1/120 (0%)
Query 43 MRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFKVT 102
M+Q V++ F+P+ + R+ E I+ +PLN++ + ++ L++ + + L ++ + T
Sbjct 133 MKQQVVELARKTFRPKFMNRIDEYIVSQPLNSSEISKIVELQMRQVKKRLEQNKINLEYT 192
Query 103 DSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVLARSNTNPNQL 162
A+ + + G R + + +EK +K + ++ G + D +L ++ N+L
Sbjct 193 KEAVDLLAQLGFDPNNGARPVKQMIEKLVKKEITLKVLKGDF-AEDGTILIDADQPNNKL 251
> ECU11g1420
Length=851
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 24/107 (22%), Positives = 54/107 (50%), Gaps = 0/107 (0%)
Query 34 RRPGDILKRMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLA 93
R G++ R+ + + VL F + R+ E++ F L+ + +L ++ L + L
Sbjct 711 RSRGELNDDERKKIEEIVLNRFGAPFVNRIDEVLYFNNLDRENMHRILEYQMGYLEKKLR 770
Query 94 AKGVDFKVTDSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLI 140
+G+ FKV+D+ ++ + S YG R++ +++ K + +L+
Sbjct 771 GRGITFKVSDAVKEDMVTKVLSSIYGARQLRRFIRSSFKRALTKVLL 817
> YLL026w
Length=908
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 23/102 (22%), Positives = 50/102 (49%), Gaps = 2/102 (1%)
Query 47 VMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGLTRSLAAKGVDFK--VTDS 104
VM V +F+P+ + R+S I+IF L+ A+ ++ ++L + +K +T
Sbjct 750 VMGAVRQHFRPEFLNRISSIVIFNKLSRKAIHKIVDIRLKEIEERFEQNDKHYKLNLTQE 809
Query 105 AMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKS 146
A F+ + S G R + + ++ I ++A ++ ++K
Sbjct 810 AKDFLAKYGYSDDMGARPLNRLIQNEILNKLALRILKNEIKD 851
> Hs18584739
Length=224
Score = 33.1 bits (74), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Query 2 NEQFPRGSGKQQQRGLVSGGAVRCRFYSAAANRRPGDILKRMRQSVMKEV 51
NE FP+GSGK L S G V+ F AA R + + +R S+ K +
Sbjct 81 NEPFPQGSGKS----LPSQGFVKFSFRQTAARRHMCNTVWALRSSIFKSM 126
> At5g13680
Length=1319
Score = 32.7 bits (73), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 114 SSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVL 152
S +K ++GKYL +Y+ R LL++ K+KS +R+V+
Sbjct 1120 SEFKESIEKVGKYLTRYLAVRQRRLLLAAKLKSEERSVV 1158
> 7290685
Length=1399
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 103 DSAMTFILERASSYKYGGRRMGKYLEKYIKPRVAPLLISGKVKSGDRAVLARSNTN 158
D A+TF+L + Y + +L +Y RVA + +VK+G + + A ++T+
Sbjct 272 DDAVTFLLPQHPLYPLSRLHVPVFLHQYPDQRVAAFTVRARVKAGLKILGATASTD 327
> At1g42705
Length=723
Score = 29.6 bits (65), Expect = 4.3, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 170 DEAGNCRPGTKYGTKLVVQEVKTQEDGDDDE 200
D A C+P K T+ V + T++DGDDD+
Sbjct 42 DSANPCKPKKKLQTRSWVWDHFTRKDGDDDQ 72
> SPBC119.17
Length=882
Score = 28.9 bits (63), Expect = 7.4, Method: Composition-based stats.
Identities = 25/98 (25%), Positives = 40/98 (40%), Gaps = 5/98 (5%)
Query 73 NTAALKGVLSLKLAGLTRSLAAKGVDFKVTDSAMTFILERASSYKYGGRRMGKYL----- 127
NT L +L ++G+T +A KG F SA + + + + GG K L
Sbjct 677 NTDKLAIMLKTSVSGITDGIAEKGHSFAKVSSASGLTEKTSITEQLGGLTQVKLLSQLSR 736
Query 128 EKYIKPRVAPLLISGKVKSGDRAVLARSNTNPNQLNLI 165
E+ P V L ++ G A N +P Q ++
Sbjct 737 EESFGPLVEKLTAIREILRGTSGFKAAINASPTQHEVV 774
> CE17543
Length=818
Score = 28.5 bits (62), Expect = 9.4, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 38/72 (52%), Gaps = 10/72 (13%)
Query 32 ANRRPGDILKRMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGL--- 88
AN+R + ++M QS MK YF +GR SE++I+E A+K ++ L G
Sbjct 127 ANQRRDENNEKMIQSKMK----YFLVLKVGRSSEVMIYERKLHDAVKYLVERFLGGRGIL 182
Query 89 ---TRSLAAKGV 97
T S+ ++G+
Sbjct 183 KVGTLSIGSRGI 194
> CE17542B
Length=818
Score = 28.5 bits (62), Expect = 9.6, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 38/72 (52%), Gaps = 10/72 (13%)
Query 32 ANRRPGDILKRMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGL--- 88
AN+R + ++M QS MK YF +GR SE++I+E A+K ++ L G
Sbjct 127 ANQRRDENNEKMIQSKMK----YFLVLKVGRSSEVMIYERKLHDAVKYLVERFLGGRGIL 182
Query 89 ---TRSLAAKGV 97
T S+ ++G+
Sbjct 183 KVGTLSIGSRGI 194
> CE17542A
Length=818
Score = 28.5 bits (62), Expect = 9.6, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 38/72 (52%), Gaps = 10/72 (13%)
Query 32 ANRRPGDILKRMRQSVMKEVLGYFKPQVIGRMSEIIIFEPLNTAALKGVLSLKLAGL--- 88
AN+R + ++M QS MK YF +GR SE++I+E A+K ++ L G
Sbjct 127 ANQRRDENNEKMIQSKMK----YFLVLKVGRSSEVMIYERKLHDAVKYLVERFLGGRGIL 182
Query 89 ---TRSLAAKGV 97
T S+ ++G+
Sbjct 183 KVGTLSIGSRGI 194
Lambda K H
0.317 0.136 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3586972600
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40