bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0396_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
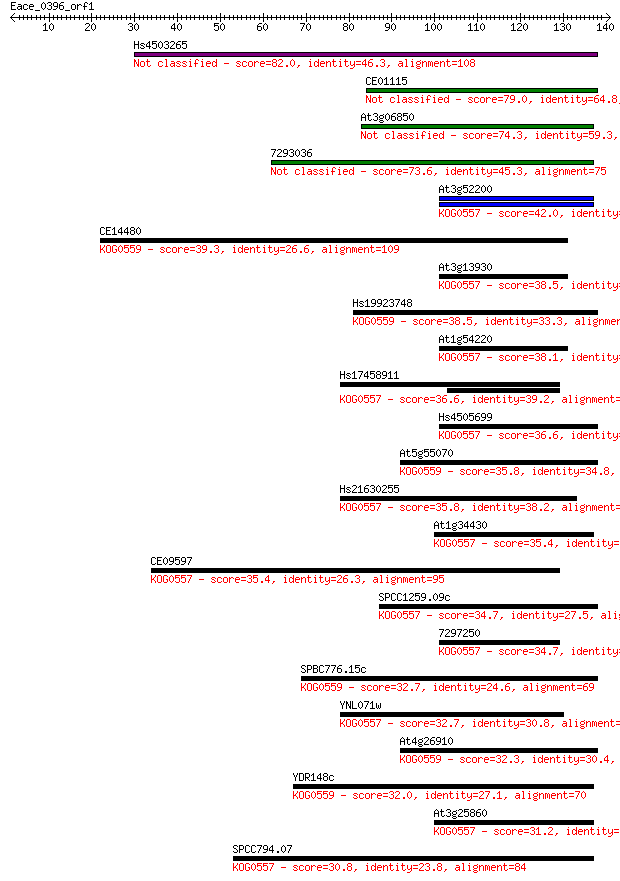
Hs4503265 82.0 3e-16
CE01115 79.0 3e-15
At3g06850 74.3 7e-14
7293036 73.6 1e-13
At3g52200 42.0 3e-04
CE14480 39.3 0.002
At3g13930 38.5 0.004
Hs19923748 38.5 0.004
At1g54220 38.1 0.005
Hs17458911 36.6 0.016
Hs4505699 36.6 0.017
At5g55070 35.8 0.024
Hs21630255 35.8 0.029
At1g34430 35.4 0.031
CE09597 35.4 0.032
SPCC1259.09c 34.7 0.052
7297250 34.7 0.065
SPBC776.15c 32.7 0.20
YNL071w 32.7 0.20
At4g26910 32.3 0.27
YDR148c 32.0 0.33
At3g25860 31.2 0.57
SPCC794.07 30.8 0.95
> Hs4503265
Length=482
Score = 82.0 bits (201), Expect = 3e-16, Method: Composition-based stats.
Identities = 50/121 (41%), Positives = 62/121 (51%), Gaps = 17/121 (14%)
Query 30 MGRVAAATSYSSNAGGLCTFLAPVAVAGVRVPRQG---LFKTPSARGIFSISQPRH---- 82
M V ++S NAG L V V + F PS F S P H
Sbjct 1 MAAVRMLRTWSRNAGKLICVRYFQTCGNVHVLKPNYVCFFGYPS----FKYSHPHHFLKT 56
Query 83 ------GIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPG 136
+V FKL+DIGEGI V + +WY KEGDT+ + + +CEVQSDKA+V ITSRY G
Sbjct 57 TAALRGQVVQFKLSDIGEGIREVTVKEWYVKEGDTVSQFDSICEVQSDKASVTITSRYDG 116
Query 137 V 137
V
Sbjct 117 V 117
> CE01115
Length=448
Score = 79.0 bits (193), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/54 (64%), Positives = 45/54 (83%), Gaps = 0/54 (0%)
Query 84 IVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPGV 137
+V FKL+DIGEGIA V++ +WY KEGDTI + ++VCEVQSDKAAV I+ RY G+
Sbjct 30 VVQFKLSDIGEGIAEVQVKEWYVKEGDTISQFDKVCEVQSDKAAVTISCRYDGI 83
> At3g06850
Length=483
Score = 74.3 bits (181), Expect = 7e-14, Method: Composition-based stats.
Identities = 32/54 (59%), Positives = 41/54 (75%), Gaps = 0/54 (0%)
Query 83 GIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPG 136
G++ LA GEGIA EL KW+ KEGD++EE + +CEVQSDKA +EITSR+ G
Sbjct 74 GLIDVPLAQTGEGIAECELLKWFVKEGDSVEEFQPLCEVQSDKATIEITSRFKG 127
> 7293036
Length=462
Score = 73.6 bits (179), Expect = 1e-13, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 62 RQGLFKTPSARGIFSISQPRHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEV 121
R + + + R ++ V F L+DIGEGI V + +W+ KEGDT+E+ + +CEV
Sbjct 16 RNCISQRAALRRCLHVTSSLDKTVSFNLSDIGEGIREVTVKEWFVKEGDTVEQFDNLCEV 75
Query 122 QSDKAAVEITSRYPG 136
QSDKA+V ITSRY G
Sbjct 76 QSDKASVTITSRYDG 90
> At3g52200
Length=637
Score = 42.0 bits (97), Expect = 3e-04, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 101 LTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPG 136
+ KW KKEGD +E + +CE+++DKA VE S+ G
Sbjct 102 VVKWMKKEGDKVEVGDVLCEIETDKATVEFESQEEG 137
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 16/36 (44%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 101 LTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPG 136
+ KW+KKEGD IE + + E+++DKA +E S G
Sbjct 229 IAKWWKKEGDKIEVGDVIGEIETDKATLEFESLEEG 264
> CE14480
Length=452
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 52/114 (45%), Gaps = 15/114 (13%)
Query 22 RMACVARYMGR-----VAAATSYSSNAGGLCTFLAPVAVAGVRVPRQGLFKTPSARGIFS 76
R V R++ R V AA+S + + L P+ V VR+ F + R
Sbjct 5 RAVSVHRFLSRAARQSVTAASSAQPSLQAKTSLLEPL-VQNVRITSSANFHMSAVR---- 59
Query 77 ISQPRHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEI 130
++ + E I+ ++ +W K++GD + E E V E+++DK +VE+
Sbjct 60 ----MSDVITVEGPAFAESISEGDI-RWLKQKGDHVNEDELVAEIETDKTSVEV 108
> At3g13930
Length=508
Score = 38.5 bits (88), Expect = 0.004, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 101 LTKWYKKEGDTIEEMEEVCEVQSDKAAVEI 130
+ +W KKEGD + E +CEV++DKA VE+
Sbjct 102 IARWLKKEGDKVAPGEVLCEVETDKATVEM 131
> Hs19923748
Length=453
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 32/57 (56%), Gaps = 1/57 (1%)
Query 81 RHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPGV 137
+ +V K E + ++ +W K GDT+ E E VCE+++DK +V++ S GV
Sbjct 67 KDDLVTVKTPAFAESVTEGDV-RWEKAVGDTVAEDEVVCEIETDKTSVQVPSPANGV 122
> At1g54220
Length=424
Score = 38.1 bits (87), Expect = 0.005, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 101 LTKWYKKEGDTIEEMEEVCEVQSDKAAVEI 130
+ +W KKEGD + E +CEV++DKA VE+
Sbjct 13 IARWLKKEGDKVAPGEVLCEVETDKATVEM 42
> Hs17458911
Length=647
Score = 36.6 bits (83), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 78 SQPRHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAV 128
S P H V L + + A + +W KKEGD I E + + EV++DKA V
Sbjct 87 SLPPHQKV--PLPSLSPTMQAGTIARWEKKEGDQINEGDLIAEVETDKATV 135
Score = 27.3 bits (59), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 9/26 (34%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 103 KWYKKEGDTIEEMEEVCEVQSDKAAV 128
+W KK G+ + E + + E+++DKA +
Sbjct 237 RWEKKVGEKLSEGDLLAEIETDKATI 262
> Hs4505699
Length=501
Score = 36.6 bits (83), Expect = 0.017, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 101 LTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPGV 137
+ KW KKEG+ + + +CE+++DKA V + + G+
Sbjct 73 IVKWLKKEGEAVSAGDALCEIETDKAVVTLDASDDGI 109
> At5g55070
Length=464
Score = 35.8 bits (81), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 92 IGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPGV 137
+GE I L + KK GD +E E + ++++DK ++I S GV
Sbjct 101 MGESITDGTLAAFLKKPGDRVEADEAIAQIETDKVTIDIASPASGV 146
> Hs21630255
Length=613
Score = 35.8 bits (81), Expect = 0.029, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 2/55 (3%)
Query 78 SQPRHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITS 132
S P H V L + + A + +W KKEGD I E + + EV++DKA V S
Sbjct 54 SLPPHQKV--PLPSLSPTMQAGTIARWEKKEGDKINEGDLIAEVETDKATVGFES 106
> At1g34430
Length=465
Score = 35.4 bits (80), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 100 ELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPG 136
++ W K EGD + + E V V+SDKA +++ + Y G
Sbjct 55 KIVSWVKSEGDKLNKGESVVVVESDKADMDVETFYDG 91
> CE09597
Length=507
Score = 35.4 bits (80), Expect = 0.032, Method: Composition-based stats.
Identities = 25/95 (26%), Positives = 42/95 (44%), Gaps = 8/95 (8%)
Query 34 AAATSYSSNAGGLCTFLAPVAVAGVRVPRQGLFKTPSARGIFSISQPRHGIVCFKLADIG 93
A +T ++ + GL V + P F R S + P+H V L +
Sbjct 35 ALSTGAAAKSSGL------VGQVARQYPNAAAFSIKQVRLYSSGNLPKHNRVA--LPALS 86
Query 94 EGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAV 128
+ + W KKEGD + E + +CE+++DKA +
Sbjct 87 PTMELGTVVSWQKKEGDQLSEGDLLCEIETDKATM 121
> SPCC1259.09c
Length=456
Score = 34.7 bits (78), Expect = 0.052, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 87 FKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPGV 137
F++ + + +TKW+ KEGD+ + + + EV++DKA +++ + G+
Sbjct 38 FRMPALSPTMEEGNITKWHFKEGDSFKSGDILLEVETDKATMDVEVQDNGI 88
> 7297250
Length=512
Score = 34.7 bits (78), Expect = 0.065, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 20/28 (71%), Gaps = 0/28 (0%)
Query 101 LTKWYKKEGDTIEEMEEVCEVQSDKAAV 128
+ W KKEGD + E + +CE+++DKA +
Sbjct 97 IVSWEKKEGDKLNEGDLLCEIETDKATM 124
> SPBC776.15c
Length=452
Score = 32.7 bits (73), Expect = 0.20, Method: Composition-based stats.
Identities = 17/69 (24%), Positives = 33/69 (47%), Gaps = 0/69 (0%)
Query 69 PSARGIFSISQPRHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAV 128
P ++ + ++ R+ K E I L +W K+ G+ + + EE+ V++DK
Sbjct 27 PVSKSMANVLWARYASTRIKTPPFPESITEGTLAQWLKQPGEYVNKDEEIASVETDKIDA 86
Query 129 EITSRYPGV 137
+T+ GV
Sbjct 87 PVTAPDAGV 95
> YNL071w
Length=482
Score = 32.7 bits (73), Expect = 0.20, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 78 SQPRHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVE 129
S P H I+ + + + L W KKEGD + E + E+++DKA ++
Sbjct 30 SYPEHTII--GMPALSPTMTQGNLAAWTKKEGDQLSPGEVIAEIETDKAQMD 79
> At4g26910
Length=511
Score = 32.3 bits (72), Expect = 0.27, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 92 IGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPGV 137
+GE I L + KK G+ ++ E + ++++DK ++I S GV
Sbjct 128 MGESITDGTLATFLKKPGERVQADEAIAQIETDKVTIDIASPASGV 173
> YDR148c
Length=463
Score = 32.0 bits (71), Expect = 0.33, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 33/70 (47%), Gaps = 0/70 (0%)
Query 67 KTPSARGIFSISQPRHGIVCFKLADIGEGIAAVELTKWYKKEGDTIEEMEEVCEVQSDKA 126
K A FSI+ R ++ + E + L ++ K GD I+E E + +++DK
Sbjct 56 KISVAANPFSITSNRFKSTSIEVPPMAESLTEGSLKEYTKNVGDFIKEDELLATIETDKI 115
Query 127 AVEITSRYPG 136
+E+ S G
Sbjct 116 DIEVNSPVSG 125
> At3g25860
Length=480
Score = 31.2 bits (69), Expect = 0.57, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 100 ELTKWYKKEGDTIEEMEEVCEVQSDKAAVEITSRYPG 136
++ W K EG+ + + E V V+SDKA +++ + Y G
Sbjct 71 KIVSWIKTEGEKLAKGESVVVVESDKADMDVETFYDG 107
> SPCC794.07
Length=483
Score = 30.8 bits (68), Expect = 0.95, Method: Composition-based stats.
Identities = 20/86 (23%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Query 53 VAVAGVRVPRQGLFKTPS--ARGIFSISQPRHGIVCFKLADIGEGIAAVELTKWYKKEGD 110
V+ V+V + L+ + AR + + P H ++ + + + + + KK GD
Sbjct 22 VSNGKVQVKKSALYPVMAKLARTYATKNYPAHTVI--NMPALSPTMTTGNIGAFQKKIGD 79
Query 111 TIEEMEEVCEVQSDKAAVEITSRYPG 136
IE + +CE+++DKA ++ + G
Sbjct 80 KIEPGDVLCEIETDKAQIDFEQQDEG 105
Lambda K H
0.322 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1571531578
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40