bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0407_orf1
Length=160
Score E
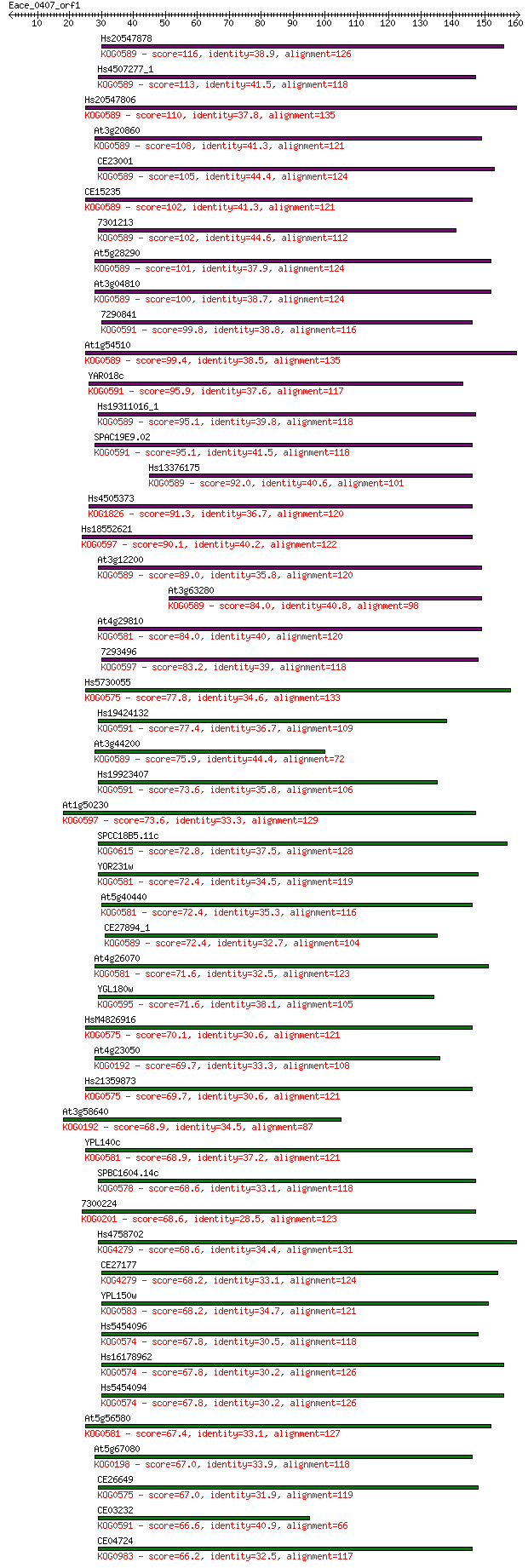
Sequences producing significant alignments: (Bits) Value
Hs20547878 116 2e-26
Hs4507277_1 113 1e-25
Hs20547806 110 1e-24
At3g20860 108 3e-24
CE23001 105 5e-23
CE15235 102 4e-22
7301213 102 4e-22
At5g28290 101 5e-22
At3g04810 100 1e-21
7290841 99.8 2e-21
At1g54510 99.4 2e-21
YAR018c 95.9 3e-20
Hs19311016_1 95.1 5e-20
SPAC19E9.02 95.1 6e-20
Hs13376175 92.0 4e-19
Hs4505373 91.3 7e-19
Hs18552621 90.1 2e-18
At3g12200 89.0 4e-18
At3g63280 84.0 1e-16
At4g29810 84.0 1e-16
7293496 83.2 2e-16
Hs5730055 77.8 8e-15
Hs19424132 77.4 1e-14
At3g44200 75.9 3e-14
Hs19923407 73.6 1e-13
At1g50230 73.6 2e-13
SPCC18B5.11c 72.8 3e-13
YOR231w 72.4 3e-13
At5g40440 72.4 4e-13
CE27894_1 72.4 4e-13
At4g26070 71.6 5e-13
YGL180w 71.6 6e-13
HsM4826916 70.1 2e-12
At4g23050 69.7 2e-12
Hs21359873 69.7 2e-12
At3g58640 68.9 4e-12
YPL140c 68.9 4e-12
SPBC1604.14c 68.6 5e-12
7300224 68.6 5e-12
Hs4758702 68.6 6e-12
CE27177 68.2 6e-12
YPL150w 68.2 7e-12
Hs5454096 67.8 8e-12
Hs16178962 67.8 9e-12
Hs5454094 67.8 9e-12
At5g56580 67.4 1e-11
At5g67080 67.0 1e-11
CE26649 67.0 1e-11
CE03232 66.6 2e-11
CE04724 66.2 2e-11
> Hs20547878
Length=758
Score = 116 bits (291), Expect = 2e-26, Method: Composition-based stats.
Identities = 49/126 (38%), Positives = 82/126 (65%), Gaps = 0/126 (0%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNA 89
K+GDFGI++V++ ++ +A T +GTP Y+SPE+ +++PY K+D+W+LGCVLYELCT ++
Sbjct 120 KLGDFGIARVLNNSMELARTCIGTPYYLSPEICQNKPYNNKTDIWSLGCVLYELCTLKHP 179
Query 90 FAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQAFA 149
F ++ LV I A PIS ++ L L++ + + P RP+ +L+ PFL+
Sbjct 180 FEGNNLQQLVLKICQAHFAPISPGFSRELHSLISQLFQVSPRDRPSINSILKRPFLENLI 239
Query 150 NRHARP 155
++ P
Sbjct 240 PKYLTP 245
> Hs4507277_1
Length=292
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 49/118 (41%), Positives = 77/118 (65%), Gaps = 0/118 (0%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+K+GD GI++V+ +A+T +GTP YMSPE+ ++PY KSDVWALGC +YE+ T ++
Sbjct 145 IKVGDLGIARVLENHCDMASTLIGTPYYMSPELFSNKPYNYKSDVWALGCCVYEMATLKH 204
Query 89 AFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQ 146
AF A +LV+ I + + Y+P L+ L+ +ML K P RP+ +LR P+++
Sbjct 205 AFNAKDMNSLVYRIIEGKLPAMPRDYSPELAELIRTMLSKRPEERPSVRSILRQPYIK 262
> Hs20547806
Length=489
Score = 110 bits (275), Expect = 1e-24, Method: Composition-based stats.
Identities = 51/135 (37%), Positives = 81/135 (60%), Gaps = 5/135 (3%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
Q +K+GDFG ++++S + A T VGTP Y+ PE+ E+ PY KSD+W+LGC+LYELC
Sbjct 137 QNGKVKLGDFGSARLLSNPMAFACTYVGTPYYVPPEIWENLPYNNKSDIWSLGCILYELC 196
Query 85 TYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPF 144
T ++ F A+S+ L+ + I P+ + Y+ L L+ M +++P+ RP+ T LL
Sbjct 197 TLKHPFQANSWKNLILKVCQGCISPLPSHYSYELQFLVKQMFKRNPSHRPSATTLLSRGI 256
Query 145 LQAFANRHARPPCLP 159
+ + CLP
Sbjct 257 VARLVQK-----CLP 266
> At3g20860
Length=427
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 50/121 (41%), Positives = 77/121 (63%), Gaps = 1/121 (0%)
Query 28 SLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYR 87
+++GDFG++K++ +A++ VGTP YM PE+L PY KSD+W+LGC ++E+ ++
Sbjct 152 EVRLGDFGLAKLLGKD-DLASSMVGTPNYMCPELLADIPYGYKSDIWSLGCCMFEVAAHQ 210
Query 88 NAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQA 147
AF A AL+ I + + P+ +Y+ L RL+ SML K+P RPT LLR P LQ
Sbjct 211 PAFKAPDMAALINKINRSSLSPLPVMYSSSLKRLIKSMLRKNPEHRPTAAELLRHPHLQP 270
Query 148 F 148
+
Sbjct 271 Y 271
> CE23001
Length=579
Score = 105 bits (261), Expect = 5e-23, Method: Composition-based stats.
Identities = 55/126 (43%), Positives = 75/126 (59%), Gaps = 4/126 (3%)
Query 29 LKIGDFGISKVM-SATLGV-ATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTY 86
+KIGDFGISK+M + TL A T VGTP Y+SPEM Y KSD+WALGC+LYE+C
Sbjct 308 VKIGDFGISKIMGTETLAQGAKTVVGTPYYISPEMCSGVSYNEKSDMWALGCILYEMCCL 367
Query 87 RNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQ 146
+ AF D+ ALV I P+ Y+ + ++ +L+ DP +RP+ L+ L+
Sbjct 368 KKAFEGDNLPALVNSIMTCAYTPVKGPYSAEMKMVIRELLQLDPQKRPSAPQALK--MLR 425
Query 147 AFANRH 152
NRH
Sbjct 426 PSENRH 431
> CE15235
Length=357
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 50/121 (41%), Positives = 73/121 (60%), Gaps = 1/121 (0%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
+K LK+ DFGISK + T A+T +GTP Y+SPE+ ESRPY KSD+W+LGCVLYEL
Sbjct 148 RKTVLKLSDFGISKEL-GTKSAASTVIGTPNYLSPEICESRPYNQKSDMWSLGCVLYELL 206
Query 85 TYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPF 144
AF ++ A+V I + P+ + + L+ ++L+ +RP + LL DP
Sbjct 207 QLERAFDGENLPAIVMKITRSKQNPLGDHVSNDVKMLVENLLKTHTDKRPDVSQLLSDPL 266
Query 145 L 145
+
Sbjct 267 V 267
> 7301213
Length=841
Score = 102 bits (253), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 50/112 (44%), Positives = 71/112 (63%), Gaps = 1/112 (0%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+KIGDFGISK+M+ + A T +GTP Y SPEM E + Y KSD+WALGC+L E+C +
Sbjct 244 VKIGDFGISKIMNTKIH-AQTVLGTPYYFSPEMCEGKEYDNKSDIWALGCILGEMCCLKK 302
Query 89 AFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLL 140
FAA + LV I P+ + YT L L++++L+ + RRPT + +L
Sbjct 303 TFAASNLSELVTKIMAGNYTPVPSGYTSGLRSLMSNLLQVEAPRRPTASEVL 354
> At5g28290
Length=568
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 47/124 (37%), Positives = 76/124 (61%), Gaps = 1/124 (0%)
Query 28 SLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYR 87
+++GDFG++K++++ +A++ VGTP YM PE+L PY KSD+W+LGC +YE+ +
Sbjct 142 DIRLGDFGLAKILTSD-DLASSVVGTPSYMCPELLADIPYGSKSDIWSLGCCMYEMTALK 200
Query 88 NAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQA 147
AF A L+ I + + P+ Y+ L+ SML K+P RP+ + LLR P LQ
Sbjct 201 PAFKAFDMQGLINRINRSIVAPLPAQYSTAFRSLVKSMLRKNPELRPSASDLLRQPLLQP 260
Query 148 FANR 151
+ +
Sbjct 261 YVQK 264
> At3g04810
Length=606
Score = 100 bits (250), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 48/124 (38%), Positives = 75/124 (60%), Gaps = 1/124 (0%)
Query 28 SLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYR 87
+++GDFG++KV+++ +A++ VGTP YM PE+L PY KSD+W+LGC +YE+ +
Sbjct 142 DIRLGDFGLAKVLTSD-DLASSVVGTPSYMCPELLADIPYGSKSDIWSLGCCMYEMTAMK 200
Query 88 NAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQA 147
AF A L+ I + + P+ Y+ L+ SML K+P RP+ LLR P LQ
Sbjct 201 PAFKAFDMQGLINRINRSIVPPLPAQYSAAFRGLVKSMLRKNPELRPSAAELLRQPLLQP 260
Query 148 FANR 151
+ +
Sbjct 261 YIQK 264
> 7290841
Length=735
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 68/116 (58%), Gaps = 0/116 (0%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNA 89
K+GDFG+++++ A + VGTP YMSPE+++ R Y KSDVWA+GC++YE+C R
Sbjct 166 KLGDFGLARMLRRDQSFAASFVGTPHYMSPELVKGRKYDRKSDVWAVGCLVYEMCALRPP 225
Query 90 FAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFL 145
F +F L IA I +Y+ L ++A ML D +RP ++R P +
Sbjct 226 FRGRAFDQLSEKIAQGEFSRIPAIYSTDLQEIIAFMLAVDHEQRPGIEVIIRHPLV 281
> At1g54510
Length=612
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/145 (35%), Positives = 81/145 (55%), Gaps = 11/145 (7%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
++ +++GDFG++K++++ + ++ VGTP YM PE+L PY KSD+W+LGC +YE+
Sbjct 139 KEQDIRLGDFGLAKILTSD-DLTSSVVGTPSYMCPELLADIPYGSKSDIWSLGCCIYEMA 197
Query 85 TYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPF 144
+ AF A AL+ I + P+ Y+ L+ SML K+P RP+ + LLR P
Sbjct 198 YLKPAFKAFDMQALINKINKTIVSPLPAKYSGPFRGLVKSMLRKNPEVRPSASDLLRHPH 257
Query 145 LQAFA----------NRHARPPCLP 159
LQ + R PP LP
Sbjct 258 LQPYVLDVKLRLNNLRRKTLPPELP 282
> YAR018c
Length=435
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 44/117 (37%), Positives = 75/117 (64%), Gaps = 0/117 (0%)
Query 26 KVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCT 85
+V +K+GDFG++K + ++ ATT VGTP YMSPE+L +PY+ SD+W+LGCV++E+C+
Sbjct 224 QVVVKLGDFGLAKSLETSIQFATTYVGTPYYMSPEVLMDQPYSPLSDIWSLGCVIFEMCS 283
Query 86 YRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRD 142
F A +++ L I + + Y+ L+ ++ SM++ + RP+ LL+D
Sbjct 284 LHPPFQAKNYLELQTKIKNGKCDTVPEYYSRGLNAIIHSMIDVNLRTRPSTFELLQD 340
> Hs19311016_1
Length=330
Score = 95.1 bits (235), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 47/120 (39%), Positives = 75/120 (62%), Gaps = 2/120 (1%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+K+GD+G++K +++ +A T VGTP YMSPE+ + Y KSD+WA+GCV++EL T +
Sbjct 190 IKLGDYGLAKKLNSEYSMAETLVGTPYYMSPELCQGVKYNFKSDIWAVGCVIFELLTLKR 249
Query 89 AFAADSFIALVWMI--AFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQ 146
F A + + L I +E S+ Y+ L +++ S L++DP +RPT LL P L+
Sbjct 250 TFDATNPLNLCVKIVQGIRAMEVDSSQYSLELIQMVHSCLDQDPEQRPTADELLDRPLLR 309
> SPAC19E9.02
Length=722
Score = 95.1 bits (235), Expect = 6e-20, Method: Composition-based stats.
Identities = 49/118 (41%), Positives = 69/118 (58%), Gaps = 0/118 (0%)
Query 28 SLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYR 87
S+K+GDFG+SK++ T + VGTP YMSPE++ S PY+ KSDVWALGCV++E+C
Sbjct 164 SVKLGDFGLSKLLDNTRVFTQSYVGTPYYMSPEIIRSSPYSAKSDVWALGCVIFEICMLT 223
Query 88 NAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFL 145
+ F S++ L I + Y+ + L+ LE + RPT LLR P L
Sbjct 224 HPFEGRSYLELQRNICQGNLSCWDHHYSDDVFLLIRHCLEVNSDLRPTTYQLLRSPIL 281
> Hs13376175
Length=463
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 41/101 (40%), Positives = 63/101 (62%), Gaps = 0/101 (0%)
Query 45 GVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNAFAADSFIALVWMIAF 104
+ATT GTP YMSPE L+ + Y KSD+W+L C+LYE+C +AFA +F+++V I
Sbjct 5 DLATTLTGTPHYMSPEALKHQGYDTKSDIWSLACILYEMCCMNHAFAGSNFLSIVLKIVE 64
Query 105 APIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFL 145
+ Y L+ ++ SML K+P+ RP+ +L+ P+L
Sbjct 65 GDTPSLPERYPKELNAIMESMLNKNPSLRPSAIEILKIPYL 105
> Hs4505373
Length=445
Score = 91.3 bits (225), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 44/120 (36%), Positives = 65/120 (54%), Gaps = 0/120 (0%)
Query 26 KVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCT 85
K ++K+GDFG++++++ A T VGTP YMSPE + Y KSD+W+LGC+LYELC
Sbjct 152 KQNVKLGDFGLARILNHDTSFAKTFVGTPYYMSPEQMNRMSYNEKSDIWSLGCLLYELCA 211
Query 86 YRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFL 145
F A S L I I Y+ L+ ++ ML RP+ +L +P +
Sbjct 212 LMPPFTAFSQKELAGKIREGKFRRIPYRYSDELNEIITRMLNLKDYHRPSVEEILENPLI 271
> Hs18552621
Length=1315
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 49/122 (40%), Positives = 70/122 (57%), Gaps = 1/122 (0%)
Query 24 AQKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYEL 83
A+ +K+ DFG ++ MS V T+ GTP YMSPE++E RPY +D+W++GC+LYEL
Sbjct 134 AKGGGIKLCDFGFARAMSTNTMVLTSIKGTPLYMSPELVEERPYDHTADLWSVGCILYEL 193
Query 84 CTYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDP 143
F A S LV +I P+ ST+ +P L +L KDP +R + LL P
Sbjct 194 AVGTPPFYATSIFQLVSLILKDPVRWPSTI-SPCFKNFLQGLLTKDPRQRLSWPDLLYHP 252
Query 144 FL 145
F+
Sbjct 253 FI 254
> At3g12200
Length=571
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 43/120 (35%), Positives = 73/120 (60%), Gaps = 1/120 (0%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+++G++G++K+++ V+ + G M PE+LE +PY KSD+W+LGC +YE+ ++
Sbjct 158 VQLGNYGLAKLINPEKPVSMVS-GISNSMCPEVLEDQPYGYKSDIWSLGCCMYEITAHQP 216
Query 89 AFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQAF 148
AF A L+ I + + P+ VY+ L +++ ML K P RPT LLR+P LQ +
Sbjct 217 AFKAPDMAGLINKINRSLMSPLPIVYSSTLKQMIKLMLRKKPEYRPTACELLRNPSLQPY 276
> At3g63280
Length=530
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 40/98 (40%), Positives = 56/98 (57%), Gaps = 0/98 (0%)
Query 51 VGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNAFAADSFIALVWMIAFAPIEPI 110
VGTP YM PE+L PY KSD+W+LGC +YE+ ++ F A L+ I ++PI
Sbjct 139 VGTPSYMCPELLADIPYGSKSDIWSLGCCMYEMAAHKPPFKASDVQTLITKIHKLIMDPI 198
Query 111 STVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQAF 148
+Y+ L+ SML K+P RP+ LL P LQ +
Sbjct 199 PAMYSGSFRGLIKSMLRKNPELRPSANELLNHPHLQPY 236
> At4g29810
Length=363
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 48/129 (37%), Positives = 69/129 (53%), Gaps = 10/129 (7%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+KI DFG+S VM+ T G+A T VGT YMSPE + Y KSD+W+LG V+ E T +
Sbjct 206 VKITDFGVSTVMTNTAGLANTFVGTYNYMSPERIVGNKYGNKSDIWSLGLVVLECATGKF 265
Query 89 AFAADSFIALVWMIAFAPIEPI---------STVYTPHLSRLLASMLEKDPARRPTPTCL 139
+A + W F +E I S ++P LS +++ L+KDP R + L
Sbjct 266 PYAPPN-QEETWTSVFELMEAIVDQPPPALPSGNFSPELSSFISTCLQKDPNSRSSAKEL 324
Query 140 LRDPFLQAF 148
+ PFL +
Sbjct 325 MEHPFLNKY 333
> 7293496
Length=805
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 68/118 (57%), Gaps = 1/118 (0%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNA 89
K+ DFG+++ M+ V T+ GTP YM+PE+L +PY +D+W+LGC+ YE +
Sbjct 140 KLCDFGLARNMTLGTHVLTSIKGTPLYMAPELLAEQPYDHHADMWSLGCIAYESMAGQPP 199
Query 90 FAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQA 147
F A S + LV MI ++ ST+ T L +LEKDP R + T LL PF++
Sbjct 200 FCASSILHLVKMIKHEDVKWPSTL-TSECRSFLQGLLEKDPGLRISWTQLLCHPFVEG 256
> Hs5730055
Length=685
Score = 77.8 bits (190), Expect = 8e-15, Method: Composition-based stats.
Identities = 46/134 (34%), Positives = 68/134 (50%), Gaps = 2/134 (1%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
+ + LK+GDFG++ + T GTP Y+SPE+L + + +SD+WALGCV+Y +
Sbjct 215 EAMELKVGDFGLAARLEPLEHRRRTICGTPNYLSPEVLNKQGHGCESDIWALGCVMYTML 274
Query 85 TYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLR-DP 143
R F + I A S++ P L+ASML K+P RP+ ++R D
Sbjct 275 LGRPPFETTNLKETYRCIREARYTMPSSLLAP-AKHLIASMLSKNPEDRPSLDDIIRHDF 333
Query 144 FLQAFANRHARPPC 157
FLQ F C
Sbjct 334 FLQGFTPDRLSSSC 347
> Hs19424132
Length=302
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 40/112 (35%), Positives = 61/112 (54%), Gaps = 3/112 (2%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+K+GD G+ + S+ A + VGTP YMSPE + Y KSD+W+LGC+LYE+ ++
Sbjct 175 VKLGDLGLGRFFSSKTTAAHSLVGTPYYMSPERIHENGYNFKSDIWSLGCLLYEMAALQS 234
Query 89 AFAAD--SFIALVWMIAFAPIEPI-STVYTPHLSRLLASMLEKDPARRPTPT 137
F D + +L I P+ S Y+ L +L+ + DP +RP T
Sbjct 235 PFYGDKMNLYSLCKKIEQCDYPPLPSDHYSEELRQLVNMCINPDPEKRPDVT 286
> At3g44200
Length=941
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 32/72 (44%), Positives = 47/72 (65%), Gaps = 1/72 (1%)
Query 28 SLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYR 87
+++GDFG++K + A + ++ VGTP YM PE+L PY KSD+W+LGC +YE+ YR
Sbjct 146 DVRLGDFGLAKTLKAD-DLTSSVVGTPNYMCPELLADIPYGFKSDIWSLGCCIYEMAAYR 204
Query 88 NAFAADSFIALV 99
AF A L+
Sbjct 205 PAFKAFDMAGLI 216
> Hs19923407
Length=313
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/109 (34%), Positives = 59/109 (54%), Gaps = 3/109 (2%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+K+GD G+ + S+ A + VGTP YMSPE + Y KSD+W+LGC+LYE+ ++
Sbjct 186 VKLGDLGLGRFFSSETTAAHSLVGTPYYMSPERIHENGYNFKSDIWSLGCLLYEMAALQS 245
Query 89 AFAAD--SFIALVWMIAFAPIEPI-STVYTPHLSRLLASMLEKDPARRP 134
F D + +L I P+ Y+ L L++ + DP +RP
Sbjct 246 PFYGDKMNLFSLCQKIEQCDYPPLPGEHYSEKLRELVSMCICPDPHQRP 294
> At1g50230
Length=269
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 43/129 (33%), Positives = 67/129 (51%), Gaps = 3/129 (2%)
Query 18 SCCCGAAQKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALG 77
+ GA V K+ DFG ++ MS V + GTP YM+PE+++ +PY D+W+LG
Sbjct 132 NILIGAGSVV--KLCDFGFARAMSTNTVVLRSIKGTPLYMAPELVKEQPYDRTVDLWSLG 189
Query 78 CVLYELCTYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPT 137
+LYEL + F +S AL+ I P++ + T + L +L K+P R T
Sbjct 190 VILYELYVGQPPFYTNSVYALIRHIVKDPVKYPDEMST-YFESFLKGLLNKEPHSRLTWP 248
Query 138 CLLRDPFLQ 146
L PF++
Sbjct 249 ALREHPFVK 257
> SPCC18B5.11c
Length=460
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 48/138 (34%), Positives = 72/138 (52%), Gaps = 10/138 (7%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRP------YTVKSDVWALGCVLYE 82
LKI DFG++KV+ T T GT Y++PE+L+S+ Y K D+W+LGCVLY
Sbjct 308 LKISDFGLAKVIHGTGTFLETFCGTMGYLAPEVLKSKNVNLDGGYDDKVDIWSLGCVLYV 367
Query 83 LCTYRNAFAADSFIALVWMIAFA--PIEP-ISTVYTPHLSRLLASMLEKDPARRPTPTCL 139
+ T FA+ S + +I+ PIEP + + L+ MLE +P +R + +
Sbjct 368 MLTASIPFASSSQAKCIELISKGAYPIEPLLENEISEEGIDLINRMLEINPEKRISESEA 427
Query 140 LRDP-FLQAFANRHARPP 156
L+ P F + H PP
Sbjct 428 LQHPWFYTVSTHEHRTPP 445
> YOR231w
Length=508
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 41/130 (31%), Positives = 71/130 (54%), Gaps = 13/130 (10%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+K+ DFG+S A +ATT GT YM+PE ++ +PY+V SDVW+LG + E+ +
Sbjct 363 VKLCDFGVSG--EAVNSLATTFTGTSFYMAPERIQGQPYSVTSDVWSLGLTILEVANGKF 420
Query 89 AFAADSFIA-------LVWMIAFAP---IEPIST-VYTPHLSRLLASMLEKDPARRPTPT 137
+++ A L+W++ F P EP S +++P + L+KD RP+P
Sbjct 421 PCSSEKMAANIAPFELLMWILTFTPELKDEPESNIIWSPSFKSFIDYCLKKDSRERPSPR 480
Query 138 CLLRDPFLQA 147
++ P+++
Sbjct 481 QMINHPWIKG 490
> At5g40440
Length=520
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 41/118 (34%), Positives = 62/118 (52%), Gaps = 2/118 (1%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNA 89
KI DFGIS + ++ + T VGT YMSPE + + Y+ +D+W+LG L+E T
Sbjct 222 KITDFGISAGLENSMAMCATFVGTVTYMSPERIRNDSYSYPADIWSLGLALFECGTGEFP 281
Query 90 FAADSF-IALVWMIAFAPI-EPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFL 145
+ A+ + L+ I P P ++P + + L+KDP RPT LL PF+
Sbjct 282 YIANEGPVNLMLQILDDPSPTPPKQEFSPEFCSFIDACLQKDPDARPTADQLLSHPFI 339
> CE27894_1
Length=1011
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 34/104 (32%), Positives = 58/104 (55%), Gaps = 2/104 (1%)
Query 31 IGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNAF 90
I DFG++K +A GT Y PE++++ PY K+D+W+ GC +YE+C + F
Sbjct 585 ITDFGLAKQKGPEY--LKSAAGTIIYSCPEIVQNLPYGEKADIWSFGCCIYEMCQLQPVF 642
Query 91 AADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRP 134
+ + + L I A +P++ +++ L L+ S L DP+ RP
Sbjct 643 HSTNMLTLAMQIVEAKYDPLNEMWSDDLRFLITSCLAPDPSARP 686
> At4g26070
Length=354
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 40/132 (30%), Positives = 68/132 (51%), Gaps = 10/132 (7%)
Query 28 SLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYR 87
+KI DFG+SK++++T +A + VGT YMSPE + Y+ KSD+W+LG VL E T +
Sbjct 203 EVKITDFGVSKILTSTSSLANSFVGTYPYMSPERISGSLYSNKSDIWSLGLVLLECATGK 262
Query 88 NAFAADSFIALVWMIAFAPIEPI---------STVYTPHLSRLLASMLEKDPARRPTPTC 138
+ W + ++ I S +++P ++ ++KDP R +
Sbjct 263 FPYTPPEH-KKGWSSVYELVDAIVENPPPCAPSNLFSPEFCSFISQCVQKDPRDRKSAKE 321
Query 139 LLRDPFLQAFAN 150
LL F++ F +
Sbjct 322 LLEHKFVKMFED 333
> YGL180w
Length=897
Score = 71.6 bits (174), Expect = 6e-13, Method: Composition-based stats.
Identities = 40/113 (35%), Positives = 62/113 (54%), Gaps = 14/113 (12%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
LKI DFG ++ + T +A T G+P YM+PE+L + Y K+D+W++G V++E+C
Sbjct 207 LKIADFGFARFLPNT-SLAETLCGSPLYMAPEILNYQKYNAKADLWSVGTVVFEMCCGTP 265
Query 89 AFAADSFIALVWMIAFAPIEPISTVYT--------PHLSRLLASMLEKDPARR 133
F A + + L F I+ + V T P L L+ S+L DPA+R
Sbjct 266 PFRASNHLEL-----FKKIKRANDVITFPSYCNIEPELKELICSLLTFDPAQR 313
> HsM4826916
Length=603
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 61/121 (50%), Gaps = 1/121 (0%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
+ + +KIGDFG++ + T GTP Y++PE+L + ++ + DVW++GC++Y L
Sbjct 186 EDLEVKIGDFGLATKVEYDGERKKTLCGTPNYIAPEVLSKKEHSFEVDVWSIGCIMYTLL 245
Query 85 TYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPF 144
+ F S + ++ I P + L+ ML+ DP RPT LL D F
Sbjct 246 VGKPPFET-SCLKETYLRIKKNEYSIPKHINPVAASLIQKMLQTDPTARPTINELLNDEF 304
Query 145 L 145
Sbjct 305 F 305
> At4g23050
Length=736
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 36/109 (33%), Positives = 62/109 (56%), Gaps = 1/109 (0%)
Query 28 SLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYR 87
++K+GDFG+SK +AT + GTPQ+M+PE+L S P K DV++ G +L+EL T
Sbjct 604 NVKVGDFGLSKWKNATFLSTKSGKGTPQWMAPEVLRSEPSNEKCDVFSFGVILWELMTTL 663
Query 88 NAFAADSFIALVWMIAFAPIE-PISTVYTPHLSRLLASMLEKDPARRPT 135
+ + I +V ++ F + P ++ ++ + DPA+RP+
Sbjct 664 VPWDRLNSIQVVGVVGFMDRRLDLPEGLNPRIASIIQDCWQTDPAKRPS 712
> Hs21359873
Length=603
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 61/121 (50%), Gaps = 1/121 (0%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
+ + +KIGDFG++ + T GTP Y++PE+L + ++ + DVW++GC++Y L
Sbjct 186 EDLEVKIGDFGLATKVEYDGERKKTLCGTPNYIAPEVLSKKGHSFEVDVWSIGCIMYTLL 245
Query 85 TYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPF 144
+ F S + ++ I P + L+ ML+ DP RPT LL D F
Sbjct 246 VGKPPFET-SCLKETYLRIKKNEYSIPKHINPVAASLIQKMLQTDPTARPTINELLNDEF 304
Query 145 L 145
Sbjct 305 F 305
> At3g58640
Length=816
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 56/87 (64%), Gaps = 0/87 (0%)
Query 18 SCCCGAAQKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALG 77
S C + K ++KI DFG+S++M+ T T + GTP++M+PE++ + P++ K D+++LG
Sbjct 686 SANCLLSNKWTVKICDFGLSRIMTGTTMRDTVSAGTPEWMAPELIRNEPFSEKCDIFSLG 745
Query 78 CVLYELCTYRNAFAADSFIALVWMIAF 104
+++ELCT + +V+ IA+
Sbjct 746 VIMWELCTLTRPWEGVPPERVVYAIAY 772
> YPL140c
Length=506
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 45/137 (32%), Positives = 70/137 (51%), Gaps = 23/137 (16%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
+K +K+ DFG+S A +A T GT YM+PE ++ +PY+V DVW+LG L E+
Sbjct 352 EKGEIKLCDFGVSG--EAVNSLAMTFTGTSFYMAPERIQGQPYSVTCDVWSLGLTLLEVA 409
Query 85 TYRNAFAADSFIALVWMIAFAPIEPISTV--YTPHL----------SRLLASM----LEK 128
R F +D V APIE ++ + ++P L S+ S L+K
Sbjct 410 GGRFPFESDKITQNV-----APIELLTMILTFSPQLKDEPELDISWSKTFRSFIDYCLKK 464
Query 129 DPARRPTPTCLLRDPFL 145
D RP+P +L+ P++
Sbjct 465 DARERPSPRQMLKHPWI 481
> SPBC1604.14c
Length=658
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 66/128 (51%), Gaps = 18/128 (14%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+K+ DFG + + + TT VGTP +M+PE++ + Y K DVW+LG + E+
Sbjct 519 IKLTDFGFCAQIDSNMTKRTTMVGTPYWMAPEVVTRKEYGFKVDVWSLGIMAIEMVEGEP 578
Query 89 AFAADSFIALVWMIAFAPIEPISTVYTPHLSR----------LLASMLEKDPARRPTPTC 138
+ ++ + +++IA T+ TP +SR L+ L +P +RP+
Sbjct 579 PYLNENPLRALYLIA--------TIGTPKISRPELLSSVFHDFLSKSLTVNPKQRPSSGE 630
Query 139 LLRDPFLQ 146
LLR PFL+
Sbjct 631 LLRHPFLK 638
> 7300224
Length=642
Score = 68.6 bits (166), Expect = 5e-12, Method: Composition-based stats.
Identities = 35/123 (28%), Positives = 65/123 (52%), Gaps = 0/123 (0%)
Query 24 AQKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYEL 83
+++ +K+ DFG++ ++ T T VGTP +M+PE+++ Y K+D+W+LG EL
Sbjct 142 SEQGDVKLADFGVAGQLTNTTSKRNTFVGTPFWMAPEVIKQSQYDAKADIWSLGITAIEL 201
Query 84 CTYRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDP 143
+ + ++++I ++ YT + + L KDP RPT LL+ P
Sbjct 202 AKGEPPNSELHPMRVLFLIPKNNPPQLTGSYTKSFKDFVEACLNKDPENRPTAKELLKYP 261
Query 144 FLQ 146
F++
Sbjct 262 FIK 264
> Hs4758702
Length=1011
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 66/135 (48%), Gaps = 6/135 (4%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRP--YTVKSDVWALGCVLYELCTY 86
LKI DFG SK ++ T GT QYM+PE+++ P Y +D+W+LGC + E+ T
Sbjct 509 LKISDFGTSKRLAGITPCTETFTGTLQYMAPEIIDQGPRGYGKAADIWSLGCTVIEMATG 568
Query 87 RNAFAA-DSFIALVWMIAFAPIE-PISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPF 144
R F S A ++ + + P+ + + L E DP R + LL DPF
Sbjct 569 RPPFHELGSPQAAMFQVGMYKVHPPMPSSLSAEAQAFLLRTFEPDPRLRASAQTLLGDPF 628
Query 145 LQAFANRHARPPCLP 159
LQ + +R P P
Sbjct 629 LQ--PGKRSRSPSSP 641
> CE27177
Length=1378
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 41/128 (32%), Positives = 62/128 (48%), Gaps = 6/128 (4%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLE--SRPYTVKSDVWALGCVLYELCTYR 87
KI DFG K ++ V T GT QYM+PE+++ R Y +D+W+ GC + E+ T R
Sbjct 806 KISDFGTCKRLAGLNPVTETFTGTLQYMAPEVIDHGQRGYGAPADIWSFGCTMVEMATGR 865
Query 88 NAFA--ADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFL 145
F + A+ + F PI T T + S + RP+ LL+DPF+
Sbjct 866 PPFVEMQNPQAAMFRVGMFKTHPPIPTEITEKCRNFIKSCFLPEACDRPSAKDLLQDPFI 925
Query 146 QAFANRHA 153
+ N H+
Sbjct 926 --YHNHHS 931
> YPL150w
Length=901
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 42/123 (34%), Positives = 63/123 (51%), Gaps = 4/123 (3%)
Query 30 KIGDFGISK-VMSATLGVATTAVGTPQYMSPEMLESRPYT-VKSDVWALGCVLYELCTYR 87
K+ DFG ++ M+ T T GT YM+PE++E R Y K D+W+LG +LY L T
Sbjct 172 KLTDFGFTRECMTKT--TLETVCGTTVYMAPELIERRTYDGFKIDIWSLGVILYTLITGY 229
Query 88 NAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQA 147
F D W I + + V L++ +L K+P RP+ + +LR PFLQ
Sbjct 230 LPFDDDDEAKTKWKIVNEEPKYDAKVIPDDARDLISRLLAKNPGERPSLSQVLRHPFLQP 289
Query 148 FAN 150
+ +
Sbjct 290 YGS 292
> Hs5454096
Length=487
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 36/123 (29%), Positives = 64/123 (52%), Gaps = 8/123 (6%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNA 89
K+ DFG++ ++ T+ T +GTP +M+PE+++ Y +D+W+LG E+ +
Sbjct 164 KLADFGVAGQLTDTMAKRNTVIGTPFWMAPEVIQEIGYNCVADIWSLGITAIEMAEGKPP 223
Query 90 FAADSFIALVWMIAFAPIEPISTVYTPHL-----SRLLASMLEKDPARRPTPTCLLRDPF 144
+A + ++MI P P T P L + + L K P +R T T LL+ PF
Sbjct 224 YADIHPMRAIFMI---PTNPPPTFRKPELWSDNFTDFVKQCLVKSPEQRATATQLLQHPF 280
Query 145 LQA 147
+++
Sbjct 281 VRS 283
> Hs16178962
Length=491
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 38/131 (29%), Positives = 68/131 (51%), Gaps = 13/131 (9%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNA 89
K+ DFG++ ++ T+ T +GTP +M+PE+++ Y +D+W+LG E+ +
Sbjct 161 KLADFGVAGQLTDTMAKRNTVIGTPFWMAPEVIQEIGYNCVADIWSLGITSIEMAEGKPP 220
Query 90 FAADSFIALVWMIAFAPIEPISTVYTPHL-----SRLLASMLEKDPARRPTPTCLLRDPF 144
+A + ++MI P P T P L + + L K+P +R T T LL+ PF
Sbjct 221 YADIHPMRAIFMI---PTNPPPTFRKPELWSDDFTDFVKKCLVKNPEQRATATQLLQHPF 277
Query 145 LQAFANRHARP 155
+ ++A+P
Sbjct 278 I-----KNAKP 283
> Hs5454094
Length=491
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 38/131 (29%), Positives = 68/131 (51%), Gaps = 13/131 (9%)
Query 30 KIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRNA 89
K+ DFG++ ++ T+ T +GTP +M+PE+++ Y +D+W+LG E+ +
Sbjct 161 KLADFGVAGQLTDTMAKRNTVIGTPFWMAPEVIQEIGYNCVADIWSLGITSIEMAEGKPP 220
Query 90 FAADSFIALVWMIAFAPIEPISTVYTPHL-----SRLLASMLEKDPARRPTPTCLLRDPF 144
+A + ++MI P P T P L + + L K+P +R T T LL+ PF
Sbjct 221 YADIHPMRAIFMI---PTNPPPTFRKPELWSDDFTDFVKKCLVKNPEQRATATQLLQHPF 277
Query 145 LQAFANRHARP 155
+ ++A+P
Sbjct 278 I-----KNAKP 283
> At5g56580
Length=356
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 67/136 (49%), Gaps = 10/136 (7%)
Query 25 QKVSLKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELC 84
K +KI DFG+S +++++G T VGT YMSPE + Y SD+W+LG + E
Sbjct 203 HKGEVKISDFGVSASLASSMGQRDTFVGTYNYMSPERISGSTYDYSSDIWSLGMSVLECA 262
Query 85 TYRNAFAAD-------SFIALVWMIAFA--PIEPISTVYTPHLSRLLASMLEKDPARRPT 135
R + SF L+ I P P S ++P +++ ++KDP R +
Sbjct 263 IGRFPYLESEDQQNPPSFYELLAAIVENPPPTAP-SDQFSPEFCSFVSACIQKDPPARAS 321
Query 136 PTCLLRDPFLQAFANR 151
LL PF++ F ++
Sbjct 322 SLDLLSHPFIKKFEDK 337
> At5g67080
Length=344
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 62/124 (50%), Gaps = 6/124 (4%)
Query 28 SLKIGDFGISKVMS--ATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCT 85
++KI DFG++K + L GTP YM+PE + Y + DVWALGCV+ E+ +
Sbjct 146 AVKIADFGLAKRIGDLTALNYGVQIRGTPLYMAPESVNDNEYGSEGDVWALGCVVVEMFS 205
Query 86 YRNAFA---ADSFIALVWMIAFAPIEP-ISTVYTPHLSRLLASMLEKDPARRPTPTCLLR 141
+ A++ +F++L+ I P I + L+ KDP +R T LL
Sbjct 206 GKTAWSLKEGSNFMSLLLRIGVGDEVPMIPEELSEQGRDFLSKCFVKDPKKRWTAEMLLN 265
Query 142 DPFL 145
PF+
Sbjct 266 HPFV 269
> CE26649
Length=648
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 38/119 (31%), Positives = 63/119 (52%), Gaps = 2/119 (1%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+KIGDFG++ ++ T GTP Y++PE+L ++ + D+WA+GC+LY L +
Sbjct 175 VKIGDFGLATTVNGD-ERKKTLCGTPNYIAPEVLNKAGHSFEVDIWAVGCILYILLFGQP 233
Query 89 AFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFLQA 147
F + S I I ++ T + L+ ML+ +P RRPT + RD F ++
Sbjct 234 PFESKSLEETYSRIRHNNYT-IPSIATQPAASLIRKMLDPEPTRRPTAKQVQRDGFFKS 291
> CE03232
Length=294
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 27/66 (40%), Positives = 41/66 (62%), Gaps = 0/66 (0%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESRPYTVKSDVWALGCVLYELCTYRN 88
+K+GD G+ + S+ A + VGTP YMSPE ++ Y KSD+W+ GC+LYE+ ++
Sbjct 164 VKLGDLGLGRFFSSKTTAAHSLVGTPYYMSPERIQESGYNFKSDLWSTGCLLYEMAALQS 223
Query 89 AFAADS 94
F D
Sbjct 224 PFYGDK 229
> CE04724
Length=505
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 38/120 (31%), Positives = 61/120 (50%), Gaps = 4/120 (3%)
Query 29 LKIGDFGISKVMSATLGVATTAVGTPQYMSPEMLESR---PYTVKSDVWALGCVLYELCT 85
+K+ DFGI+ + + + A G P YM PE L+ Y ++SDVW+ G L EL T
Sbjct 207 IKLCDFGIAGRLIESRAHSKQA-GCPLYMGPERLDPNNFDSYDIRSDVWSFGVTLVELAT 265
Query 86 YRNAFAADSFIALVWMIAFAPIEPISTVYTPHLSRLLASMLEKDPARRPTPTCLLRDPFL 145
+ +A F + ++ P ++P +L+ S L++DP RP LL+ PF+
Sbjct 266 GQYPYAGTEFDMMSKILNDEPPRLDPAKFSPDFCQLVESCLQRDPTMRPNYDMLLQHPFV 325
Lambda K H
0.322 0.132 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2172509160
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40