bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0454_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
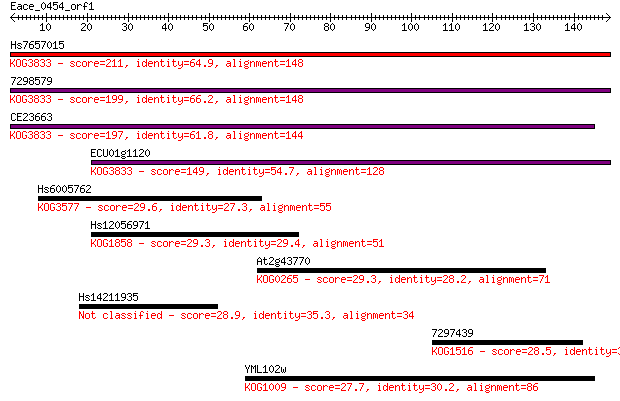
Hs7657015 211 4e-55
7298579 199 2e-51
CE23663 197 8e-51
ECU01g1120 149 1e-36
Hs6005762 29.6 2.4
Hs12056971 29.3 2.8
At2g43770 29.3 3.1
Hs14211935 28.9 3.3
7297439 28.5 4.6
YML102w 27.7 7.3
> Hs7657015
Length=505
Score = 211 bits (537), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 96/148 (64%), Positives = 118/148 (79%), Gaps = 0/148 (0%)
Query 1 DALVKMEGCMQKNKFSPIDKQLACAPIHSPEGQEYLAAMAAAANFAWVNRGAVTFLVRAA 60
DALV ME M+++K D+QLACA I SPEGQ+YL MAAA N+AWVNR ++TFL R A
Sbjct 272 DALVAMEKAMKRDKIIVNDRQLACARIASPEGQDYLKGMAAAGNYAWVNRSSMTFLTRQA 331
Query 61 FSKVFKTSPDELEMQMLYDVSHNIAKVEEHLVNGCVKELLVHRKGSTRAFPPHHPLTPVA 120
F+KVF T+PD+L++ ++YDVSHNIAKVE+H+V+G + LLVHRKGSTRAFPPHHPL V
Sbjct 332 FAKVFNTTPDDLDLHVIYDVSHNIAKVEQHVVDGKERTLLVHRKGSTRAFPPHHPLIAVD 391
Query 121 FQSIGQPVLIGGTMGSSSYILLGTSLSM 148
+Q GQPVLIGGTMG+ SY+L GT M
Sbjct 392 YQLTGQPVLIGGTMGTCSYVLTGTEQGM 419
> 7298579
Length=506
Score = 199 bits (505), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 98/148 (66%), Positives = 122/148 (82%), Gaps = 0/148 (0%)
Query 1 DALVKMEGCMQKNKFSPIDKQLACAPIHSPEGQEYLAAMAAAANFAWVNRGAVTFLVRAA 60
DALV+ME M+++K D+QLACA I+S EGQ+YL AMAAAANFAWVNR ++TFL R A
Sbjct 273 DALVQMEKAMKRDKIETNDRQLACARINSVEGQDYLKAMAAAANFAWVNRSSMTFLTRQA 332
Query 61 FSKVFKTSPDELEMQMLYDVSHNIAKVEEHLVNGCVKELLVHRKGSTRAFPPHHPLTPVA 120
F+K+F T+PD+L+M ++YDVSHNIAKVE H+V+G ++LLVHRKGSTRAFPPHHPL PV
Sbjct 333 FAKMFNTTPDDLDMHVIYDVSHNIAKVENHMVDGKERKLLVHRKGSTRAFPPHHPLIPVD 392
Query 121 FQSIGQPVLIGGTMGSSSYILLGTSLSM 148
+Q GQPVL+GGTMG+ SY+L GT M
Sbjct 393 YQLTGQPVLVGGTMGTCSYVLTGTEQGM 420
> CE23663
Length=505
Score = 197 bits (500), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 89/144 (61%), Positives = 113/144 (78%), Gaps = 0/144 (0%)
Query 1 DALVKMEGCMQKNKFSPIDKQLACAPIHSPEGQEYLAAMAAAANFAWVNRGAVTFLVRAA 60
D+LV+ME M ++ DKQLACA I+S EG+ Y + MAAAANFAWVNR +TF VR A
Sbjct 272 DSLVEMEKAMARDGIVVNDKQLACARINSVEGKNYFSGMAAAANFAWVNRSCITFCVRNA 331
Query 61 FSKVFKTSPDELEMQMLYDVSHNIAKVEEHLVNGCVKELLVHRKGSTRAFPPHHPLTPVA 120
F K F S D+++MQ++YDVSHN+AK+EEH+V+G ++L VHRKG+TRAFP HHPL PV
Sbjct 332 FQKTFGMSADDMDMQVIYDVSHNVAKMEEHMVDGRPRQLCVHRKGATRAFPAHHPLIPVD 391
Query 121 FQSIGQPVLIGGTMGSSSYILLGT 144
+Q IGQPVLIGG+MG+ SY+L GT
Sbjct 392 YQLIGQPVLIGGSMGTCSYVLTGT 415
> ECU01g1120
Length=477
Score = 149 bits (377), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 70/128 (54%), Positives = 92/128 (71%), Gaps = 4/128 (3%)
Query 21 QLACAPIHSPEGQEYLAAMAAAANFAWVNRGAVTFLVRAAFSKVFKTSPDELEMQMLYDV 80
QLAC+P +S E QEYL +M AANFA+VNR VT R AF KVF S ++++YDV
Sbjct 268 QLACSPYNSKESQEYLLSMGCAANFAFVNRAMVTEKARQAFGKVFPGS----RLELVYDV 323
Query 81 SHNIAKVEEHLVNGCVKELLVHRKGSTRAFPPHHPLTPVAFQSIGQPVLIGGTMGSSSYI 140
HNI K+E+H V+G + +VHRKG++RAFPPHHP P ++ IGQPV++GG+MG+ SYI
Sbjct 324 CHNIVKIEKHRVHGVEYDAIVHRKGASRAFPPHHPDIPWKYREIGQPVVVGGSMGTYSYI 383
Query 141 LLGTSLSM 148
L G+ SM
Sbjct 384 LCGSEKSM 391
> Hs6005762
Length=581
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 24/55 (43%), Gaps = 0/55 (0%)
Query 8 GCMQKNKFSPIDKQLACAPIHSPEGQEYLAAMAAAANFAWVNRGAVTFLVRAAFS 62
GC KF ++K +CAP+ +P Y + W+ AV +AF+
Sbjct 191 GCDNPGKFHHVEKSASCAPLCTPGVDVYWSREDKRFAVVWLAIWAVLCFFSSAFT 245
> Hs12056971
Length=1944
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 21 QLACAPIHSPEGQEYLAAMAAAANFAWVNRGAVTFLVRAAFSKVFKTSPDE 71
Q +C +HS EG++Y+A++ W + + F A+ +V SP E
Sbjct 155 QSSCINMHSIEGKDYIASLPFQVANVWPTKYGLLFERSASSHEVPPGSPRE 205
> At2g43770
Length=343
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 20/91 (21%), Positives = 34/91 (37%), Gaps = 20/91 (21%)
Query 62 SKVFKTSPDELEMQMLYDVSHNIAKVEEH--LVNGC------------------VKELLV 101
S++ SPD+ + I K+ EH VN C K +
Sbjct 109 SQIVSASPDKTVRAWDVETGKQIKKMAEHSSFVNSCCPTRRGPPLIISGSDDGTAKLWDM 168
Query 102 HRKGSTRAFPPHHPLTPVAFQSIGQPVLIGG 132
++G+ + FP + +T V+F + GG
Sbjct 169 RQRGAIQTFPDKYQITAVSFSDAADKIFTGG 199
> Hs14211935
Length=344
Score = 28.9 bits (63), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 18 IDKQLACAPIHSPEGQEYLAAMAAAANFAWVNRG 51
+++ L C +HSPE +L ++ +NF V RG
Sbjct 54 VEEGLLCRVVHSPEFNLFLDSVVFESNFIQVKRG 87
> 7297439
Length=1026
Score = 28.5 bits (62), Expect = 4.6, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 105 GSTRAFPPHHPLTPVAFQSIGQPVLIGGTMGSSSYIL 141
G F P HP + S+ P++IG T +S++I+
Sbjct 352 GDPLGFLPSHPASLATNSSLALPMIIGATKDASAFIV 388
> YML102w
Length=468
Score = 27.7 bits (60), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 42/88 (47%), Gaps = 8/88 (9%)
Query 59 AAFSKVFKTSPDELEMQMLYDVSHNIAKVEEHLV--NGCVKELLVHRKGSTRAFPPHHPL 116
AAFS VF + + +++ Y + IA E LV ++ L V G+ + P+
Sbjct 323 AAFSPVFYETCQKSVLKLPYKLVFAIATTNEVLVYDTDVLEPLCV--VGNIH----YSPI 376
Query 117 TPVAFQSIGQPVLIGGTMGSSSYILLGT 144
T +A+ G +LI T G SY+ + T
Sbjct 377 TDLAWSEDGSTLLISSTDGFCSYVSIDT 404
Lambda K H
0.320 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1821716300
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40