bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0476_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
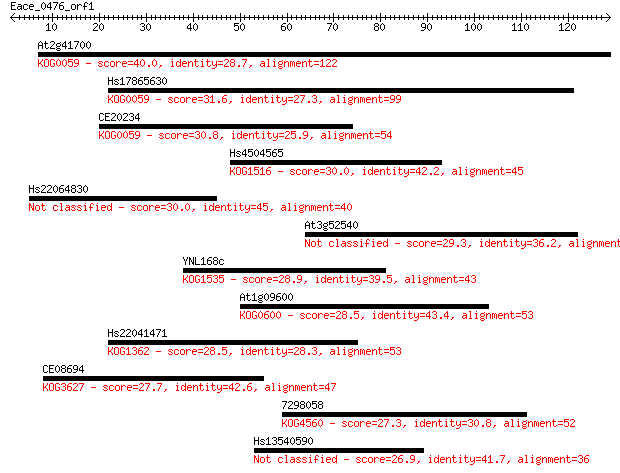
At2g41700 40.0 0.001
Hs17865630 31.6 0.34
CE20234 30.8 0.62
Hs4504565 30.0 1.2
Hs22064830 30.0 1.3
At3g52540 29.3 2.0
YNL168c 28.9 2.3
At1g09600 28.5 3.1
Hs22041471 28.5 3.2
CE08694 27.7 5.9
7298058 27.3 7.3
Hs13540590 26.9 9.6
> At2g41700
Length=1850
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 59/132 (44%), Gaps = 13/132 (9%)
Query 7 GIRSSTLNTSSLGISLADTYISLSISTIMFTCLGAYLHKIQPTSNGSPEHPLFFLSPVVS 66
G+R S + +S G+S + + + +I++ LG YL K+ P NG F S
Sbjct 373 GLRWSNIWRASSGVSFFVCLLMMLLDSILYCALGLYLDKVLPRENGVRYPWNFIFSKY-- 430
Query 67 FIRQSSRL----------CFRRRPPKNCQELQEPSESPFSRDSRGEEEDGKPILEVRDVH 116
F R+ + L F N E +P S + R +E DG+ I +VR++H
Sbjct 431 FGRKKNNLQNRIPGFETDMFPADIEVNQGEPFDPVFESISLEMRQQELDGRCI-QVRNLH 489
Query 117 KNFRTWKAKKCS 128
K + + + C+
Sbjct 490 KVYASRRGNCCA 501
> Hs17865630
Length=1642
Score = 31.6 bits (70), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 43/105 (40%), Gaps = 18/105 (17%)
Query 22 LADTYISLSISTIMFTCLGAYLHKIQPTSNGSPEHPLFFLSPVVSFIRQSSRLCFRRRPP 81
L T I L++++I + L YL ++ P G L+FL P S+ +S R +
Sbjct 396 LIITIIMLTLNSIFYVLLAVYLDQVIPGEFGLRRSSLYFLKP--SYWSKSKR-NYEELSE 452
Query 82 KNCQ------ELQEPSESPFSRDSRGEEEDGKPILEVRDVHKNFR 120
N E+ EP S F GK + + + K +R
Sbjct 453 GNVNGNISFSEIIEPVSSEFV---------GKEAIRISGIQKTYR 488
> CE20234
Length=1802
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 24/54 (44%), Gaps = 0/54 (0%)
Query 20 ISLADTYISLSISTIMFTCLGAYLHKIQPTSNGSPEHPLFFLSPVVSFIRQSSR 73
++ I+L + I+ L Y+ + P G P+ P FF+ P F S+
Sbjct 501 LTFGHALIALIVDGIIMIILTWYIEAVIPGGEGVPQKPWFFVLPSYWFPNSGSK 554
> Hs4504565
Length=559
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 25/55 (45%), Gaps = 10/55 (18%)
Query 48 PTSNGSPEHPLF---------FLSPVVSFIRQSSRLCFRRRP-PKNCQELQEPSE 92
P G P PLF L P V ++ RL F ++ P+ QEL+EP E
Sbjct 500 PNGEGLPHWPLFDQEEQYLQLNLQPAVGRALKAHRLQFWKKALPQKIQELEEPEE 554
> Hs22064830
Length=154
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 24/47 (51%), Gaps = 8/47 (17%)
Query 5 IGGIRSSTLNTSSLGISLADTY-------ISLSISTIMFTCLGAYLH 44
+GG+ L SSL I L DTY LSI + FT +G ++H
Sbjct 109 VGGLTDIVLANSSLDIVLHDTYYVVAHFHYVLSIGAV-FTIMGGFIH 154
> At3g52540
Length=282
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 7/60 (11%)
Query 64 VVSFIRQSSRLCFRRRPPKNCQELQEPSESPFSRDSRGEEEDGKPI--LEVRDVHKNFRT 121
V+ ++ S RL F RR N L+E ++ RD EEEDG + LE D + +F+
Sbjct 98 VIKGLKSSKRLIFERRGTSNSI-LEEATK----RDDHEEEEDGLMLLSLESNDPYTDFKN 152
> YNL168c
Length=259
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 23/45 (51%), Gaps = 2/45 (4%)
Query 38 CLG-AYLHKIQPTSNGSPEHPLFFLSPVVSFIR-QSSRLCFRRRP 80
C+G Y I+ +N +P+ P FFL P S + SS L RP
Sbjct 14 CIGRNYAAHIKELNNSTPKQPFFFLKPTSSIVTPLSSSLVKTTRP 58
> At1g09600
Length=714
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 30/70 (42%), Gaps = 17/70 (24%)
Query 50 SNGSPEHPLFFLSPV-----------VSFIRQSSRLCFRRRP------PKNCQELQEPSE 92
SN + E L L P+ V+ R+SSRL F+RRP N LQ+P
Sbjct 62 SNDNKEASLTLLIPIDAKKDDESEKKVNLERKSSRLVFQRRPTGIEVGANNIGTLQQPKM 121
Query 93 SPFSRDSRGE 102
+ S GE
Sbjct 122 TRICSVSNGE 131
> Hs22041471
Length=605
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 31/55 (56%), Gaps = 6/55 (10%)
Query 22 LADTYISL--SISTIMFTCLGAYLHKIQPTSNGSPEHPLFFLSPVVSFIRQSSRL 74
+A +++S+ ++ +F C L T++GS E P F +SF+++S++L
Sbjct 530 VAHSFLSVFETVLDALFLCFAVDLE----TNDGSSEKPYFMDQEFLSFVKRSNKL 580
> CE08694
Length=377
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 28/55 (50%), Gaps = 8/55 (14%)
Query 8 IRSSTLNTSSLGISLAD----TYISLSISTIMFT----CLGAYLHKIQPTSNGSP 54
I S TL ++S+ I D T+ LS+ ++ T C GAYLH P +G P
Sbjct 194 INSQTLQSTSVPIISDDDCVKTWRFLSLLSVKITGYQICAGAYLHGTAPGDSGGP 248
> 7298058
Length=1906
Score = 27.3 bits (59), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 28/56 (50%), Gaps = 4/56 (7%)
Query 59 FFLSPVVSFIRQSSRLCFRRRPPKNCQELQEPSESP----FSRDSRGEEEDGKPIL 110
FF SP+ IR+ + R PP + + P+E P + R + +E+ G P++
Sbjct 40 FFPSPLPDRIREQTWTLLLRTPPHKIEFFELPAERPLMPYYDRLNDIDEQLGLPVV 95
> Hs13540590
Length=453
Score = 26.9 bits (58), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 24/43 (55%), Gaps = 7/43 (16%)
Query 53 SPEHPLFF----LSPVVSFI---RQSSRLCFRRRPPKNCQELQ 88
SP HP F SPV++F R + F+R PP+ C++++
Sbjct 306 SPGHPAFLEDGSPSPVLAFAASPRPNHSYIFKREPPEGCEKVR 348
Lambda K H
0.317 0.133 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40