bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0478_orf1
Length=258
Score E
Sequences producing significant alignments: (Bits) Value
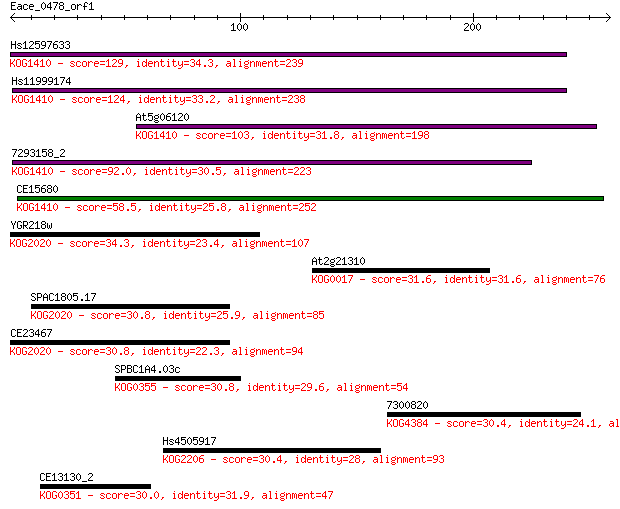
Hs12597633 129 7e-30
Hs11999174 124 1e-28
At5g06120 103 5e-22
7293158_2 92.0 1e-18
CE15680 58.5 1e-08
YGR218w 34.3 0.29
At2g21310 31.6 1.8
SPAC1805.17 30.8 2.6
CE23467 30.8 2.7
SPBC1A4.03c 30.8 3.1
7300820 30.4 3.9
Hs4505917 30.4 4.3
CE13130_2 30.0 4.6
> Hs12597633
Length=1088
Score = 129 bits (323), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 82/247 (33%), Positives = 142/247 (57%), Gaps = 15/247 (6%)
Query 1 HKVLLPLVNNPENMPRLQAVLAHSSNMQALIFASSGLMNLFTRYWGQISDKQKEETRNFL 60
K LL L+++PE + + Q +L + A + A++ L L +R + +Q+ + RN++
Sbjct 31 EKALLELIDSPECLSKCQLLLEQGTTSYAQLLAATCLSKLVSRV-SPLPVEQRMDIRNYI 89
Query 61 LNYLYQRSADLLHNAQEILGHLIRLLCRIVKLSWFED-----VDKKILEQVGLFLSASTA 115
LNY+ + A ++ LI+++ +I KL WFE V ++I+ V FL +
Sbjct 90 LNYVASQP----KLAPFVIQALIQVIAKITKLGWFEVQKDQFVFREIIADVKKFLQGTVE 145
Query 116 HWVVGLYIYTELTQEMQPKIGYH--MSRLRRVAFSFRDIALSSILKVAVKTLQQFDAGAI 173
H ++G+ I +ELTQEM + Y ++ R++A SFRD +L +L +A L++ A +
Sbjct 146 HCIIGVIILSELTQEMN-LVDYSRPSAKHRKIATSFRDTSLKDVLVLACSLLKEVFAKPL 204
Query 174 RVPNDEEETRLLKQVLQLALNCLSFDFMGTIADDTTDEQNTVMIPQGW-SEFREERFPKM 232
+ D+ + L+ QVL+L LNCL+FDF+G+ AD++ D+ TV IP W + F E +
Sbjct 205 NL-QDQCQQNLVMQVLKLVLNCLNFDFIGSSADESADDLCTVQIPTTWRTIFLEPETLDL 263
Query 233 FFSLYNA 239
FF+LY++
Sbjct 264 FFNLYHS 270
> Hs11999174
Length=1087
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 79/245 (32%), Positives = 135/245 (55%), Gaps = 12/245 (4%)
Query 2 KVLLPLVNNPENMPRLQAVLAHSSNMQALIFASSGLMNLFTRYWGQISDKQKEETRNFLL 61
K L+ N+P+ + + Q +L S+ + + A++ L L +R + +Q+ + RN++L
Sbjct 32 KALVEFTNSPDCLSKCQLLLERGSSSYSQLLAATCLTKLVSRTNNPLPLEQRIDIRNYVL 91
Query 62 NYLYQRSADLLHNAQEILGHLIRLLCRIVKLSWFE-----DVDKKILEQVGLFLSASTAH 116
NYL R A + LI+L RI KL WF+ V + + V FL S +
Sbjct 92 NYLATRP----KLATFVTQALIQLYARITKLGWFDCQKDDYVFRNAITDVTRFLQDSVEY 147
Query 117 WVVGLYIYTELTQEM-QPKIGYHMSRLRRVAFSFRDIALSSILKVAVKTLQQFDAGAIRV 175
++G+ I ++LT E+ Q + +++ R++A SFRD +L I ++ L+Q + +
Sbjct 148 CIIGVTILSQLTNEINQADTTHPLTKHRKIASSFRDSSLFDIFTLSCNLLKQASGKNLNL 207
Query 176 PNDEEETRLLKQVLQLALNCLSFDFMGTIADDTTDEQNTVMIPQGW-SEFREERFPKMFF 234
NDE + LL Q+L+L NCL+FDF+GT D+++D+ TV IP W S F + ++FF
Sbjct 208 -NDESQHGLLMQLLKLTHNCLNFDFIGTSTDESSDDLCTVQIPTSWRSAFLDSSTLQLFF 266
Query 235 SLYNA 239
LY++
Sbjct 267 DLYHS 271
> At5g06120
Length=1059
Score = 103 bits (256), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 63/201 (31%), Positives = 98/201 (48%), Gaps = 14/201 (6%)
Query 55 ETRNFLLNYLYQRSADLLHNAQEILGHLIRLLCRIVKLSWFED--VDKKILEQVGLFLSA 112
+ R +++NYL R + ++ LI+LLCR+ K W +D + E
Sbjct 79 DIRAYIVNYLATRGPKM---QSFVIASLIQLLCRLTKFGWLDDDRFRDVVKESTNFLEQG 135
Query 113 STAHWVVGLYIYTELTQEM-QPKIGYHMSRLRRVAFSFRDIALSSILKVAVKTLQQFDAG 171
S+ H+ +GL I +L QEM QP G + RRVA +FRD +L + ++A+ +L
Sbjct 136 SSDHYAIGLRILDQLVQEMNQPNPGLPSTHHRRVACNFRDQSLFQVFRIALTSLSYL--- 192
Query 172 AIRVPNDEEETRLLKQVLQLALNCLSFDFMGTIADDTTDEQNTVMIPQGWSEFREERFPK 231
++ RL + L LAL C+SFDF+GT D++T+E TV IP W E+
Sbjct 193 -----KNDAAGRLQELALSLALRCVSFDFVGTSIDESTEEFGTVQIPTSWRSVLEDSSTL 247
Query 232 MFFSLYNACWQKNPHRPRVEC 252
F Y + + +EC
Sbjct 248 QIFFDYYGSTESPLSKEALEC 268
> 7293158_2
Length=500
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 68/236 (28%), Positives = 119/236 (50%), Gaps = 21/236 (8%)
Query 2 KVLLPLVNNPENMPRLQAVLAHSSNMQALIFASSGLMNLFTRYWGQISDKQKEETRNFLL 61
K L+ V++ + +P+ Q +L + + A + A+S L T+ +S +++ + R++ L
Sbjct 26 KALVTFVSSQDALPKCQLLLQRADSSYAQLLAASTL----TKLIQGLSLQERIDIRSYAL 81
Query 62 NYLYQRSADLLHNAQEILGHLIRLLCRIVKLSWF-----EDVDKKILEQVGLFLSASTAH 116
NYL A + + ++ L+ LL ++ K WF E V + +LE V FL S H
Sbjct 82 NYL----ATVPNLQHFVVQALVSLLAKLTKYGWFDSYKEEMVFQNLLEDVKKFLQGSVEH 137
Query 117 WVVGLYIYTELTQEM----QPKIGYHMSRLRRVAFSFRDIALSSILKVAVKTLQQFDAGA 172
+G+ I ++L EM + + S++R++A SFRD L ++ L +
Sbjct 138 CTIGVQILSQLVCEMNSVVEMDVQVSFSKMRKIATSFRDQQLLETFLLSCSLLVSARDNS 197
Query 173 IRVP-NDEEETRLLKQVLQLALNCLSFDFMGTIADDTTDEQNTVM---IPQGWSEF 224
+ DE + L+ VL+L NCLSFDF+G+ D++ D+ N V P + EF
Sbjct 198 KNISFMDESQQALISHVLRLTKNCLSFDFIGSSTDESADDMNNVQGLSDPDNYHEF 253
> CE15680
Length=1078
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 65/272 (23%), Positives = 113/272 (41%), Gaps = 31/272 (11%)
Query 4 LLPLVNNPENMPRLQAVLAHSSNMQALIFASSGLMNLFTRYWGQISDKQKEETRNFLLNY 63
L L +PE + R + A + AS+ LM L I+ QK E +LL
Sbjct 30 LAELSESPECLQRCMLLFARGDYPYGPMVASTTLMKLLGGKTS-ITSVQKLELAKYLLEM 88
Query 64 LYQRSADLLHNAQEILGHLIRLLCRIVKLSW------------------FEDVDKKILEQ 105
L Q + ++ L +L R+ K W F D +++
Sbjct 89 LGQGAPQF---PPYLVTSLCQLFARLTKQEWTYQNPTENQTEDTKIEYPFRDPVDSLVKT 145
Query 106 VGLFLSASTAHWVVGLYIYTELTQEMQPKIGYH-MSRLRRVAFSFRDIALSSILKVAVKT 164
+ + + ++ + + T L +M G +++ R+ FRD L I V++
Sbjct 146 INM---DNIEESMLAVQLLTLLVADMNSASGMDSVNKHRKNLSQFRDDFLYEIFSVSLNV 202
Query 165 LQQFDAGAIRVPNDEEETRLLKQVLQLALNCLSFDFMGTIADDTTDEQNTVMIPQGW-SE 223
L + R ND + LL VL L L CL FD++G++ D+T+++ V IP W +
Sbjct 203 L---NDNVDRNLNDRQ-LGLLHAVLNLNLQCLLFDYIGSLTDETSEDNCNVQIPTAWRAS 258
Query 224 FREERFPKMFFSLYNACWQKNPHRPRVECARL 255
F + + ++ F L N Q++ + A+L
Sbjct 259 FTDGKIVQLMFKLLNVLPQESSEKVMTIIAQL 290
> YGR218w
Length=1084
Score = 34.3 bits (77), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 25/109 (22%), Positives = 49/109 (44%), Gaps = 2/109 (1%)
Query 1 HKVLLPLVNNPENMPRLQAVLAHSSNMQALIFASSGLMNLFTRYWGQISDKQKEETRNFL 60
++L +NP+ + +L S+N Q+ A S L L TR W + + + RNF+
Sbjct 35 QEILTKFQDNPDAWQKADQILQFSTNPQSKFIALSILDKLITRKWKLLPNDHRIGIRNFV 94
Query 61 LNYLYQRSAD--LLHNAQEILGHLIRLLCRIVKLSWFEDVDKKILEQVG 107
+ + D + + ++ L +I+K W ++ + I E +G
Sbjct 95 VGMIISMCQDDEVFKTQKNLINKSDLTLVQILKQEWPQNWPEFIPELIG 143
> At2g21310
Length=838
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Query 131 MQPKIGYHMSRLRRVAFSFRDIALSSILKVAVKTLQQFDA-GAIRVPNDEEETRLLKQVL 189
M +HM+ RR F D + + +K+A +T + G+IR+ ND+ T LLK V
Sbjct 307 MDTGCSFHMTP-RRDWFVEFDESQTGRVKMANQTYSEIKGIGSIRIQNDDNTTVLLKNVR 365
Query 190 QLALNCLSFDFMGTIAD 206
+ + MGT+ D
Sbjct 366 YVPSMSKNLISMGTLED 382
> SPAC1805.17
Length=966
Score = 30.8 bits (68), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 22/87 (25%), Positives = 40/87 (45%), Gaps = 2/87 (2%)
Query 10 NPENMPRLQAVLAHSSNMQALIFASSGLMNLFTRYWGQISDKQKEETRNFLLNYLYQRSA 69
+P+ + ++L S Q A S L L T W + +Q+ RN+++ + + S+
Sbjct 44 HPDAWSQAYSILEKSEYPQTKYIALSVLDKLITTRWKMLPKEQRLGIRNYIVAVMIKNSS 103
Query 70 D--LLHNAQEILGHLIRLLCRIVKLSW 94
D +L + L L L +I+K W
Sbjct 104 DETVLQQQKTFLNKLDLTLVQILKQEW 130
> CE23467
Length=1080
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 21/96 (21%), Positives = 51/96 (53%), Gaps = 2/96 (2%)
Query 1 HKVLLPLVNNPENMPRLQAVLAHSSNMQALIFASSGLMNLFTRYWGQISDKQKEETRNFL 60
+++L+ L ++ ++ A+L +S ++ FA L + W + Q+E ++++
Sbjct 46 NQILMSLKEERDSWTKVDAILQYSQLNESKYFALQILETVIQHKWKSLPQVQREGIKSYI 105
Query 61 LNYLYQRSAD--LLHNAQEILGHLIRLLCRIVKLSW 94
+ +++ S+D ++ +Q +L L +L +IVK W
Sbjct 106 ITKMFELSSDQSVMEQSQLLLHKLNLVLVQIVKQDW 141
> SPBC1A4.03c
Length=1485
Score = 30.8 bits (68), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 28/59 (47%), Gaps = 5/59 (8%)
Query 46 GQISDKQKEETRNFLLNY-----LYQRSADLLHNAQEILGHLIRLLCRIVKLSWFEDVD 99
G +S + + N+LL+ Y+R +LL E++ L L+ + K W D+D
Sbjct 1138 GDVSQDEDSDAYNYLLSMPLWSLTYERYVELLKKKDEVMAELDALIKKTPKELWLHDLD 1196
> 7300820
Length=442
Score = 30.4 bits (67), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 36/84 (42%), Gaps = 6/84 (7%)
Query 163 KTLQQFDAGAIRVPNDEEETRLLKQVLQL-ALNCLSFDFMGTIADDTTDEQNTVMIPQGW 221
K L+ D + + N E +LL V L + C D G+ +++ N + + G
Sbjct 160 KELEPADLDYLGILNQEHRAKLLTAVQLLHDIECSDVDIPGSSSENDEARLNNINMKHGA 219
Query 222 SEFREERFPKMFFSLYNACWQKNP 245
S F FP+ + C++ +P
Sbjct 220 SPFGRRHFPR-----DSGCYEGSP 238
> Hs4505917
Length=860
Score = 30.4 bits (67), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 46/95 (48%), Gaps = 4/95 (4%)
Query 67 RSADLLHNAQEILGHLIRLLCRIVKLSWFEDVDKKI--LEQVGLFLSASTAHWVVGLYIY 124
++A LL+ + L HL++L C + ++ D +I L + L + H++ LYIY
Sbjct 391 QAARLLNLGRHSLDHLLKLYCNVDSNKQYQLADWRIRPLPEEMLSYARDDTHYL--LYIY 448
Query 125 TELTQEMQPKIGYHMSRLRRVAFSFRDIALSSILK 159
++ EM + +L+ V RDI L +K
Sbjct 449 DKMRLEMWERGNGQPVQLQVVWQRSRDICLKKFIK 483
> CE13130_2
Length=873
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 25/49 (51%), Gaps = 2/49 (4%)
Query 14 MPRLQAVLAHSSNMQALIFASSG--LMNLFTRYWGQISDKQKEETRNFL 60
+PR + L +M + G +M L YW Q+ ++++EE RN L
Sbjct 734 LPRTNSDLLRIDSMTQIKVTKYGRLIMELLATYWKQVDEREEEEMRNQL 782
Lambda K H
0.324 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5400706832
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40