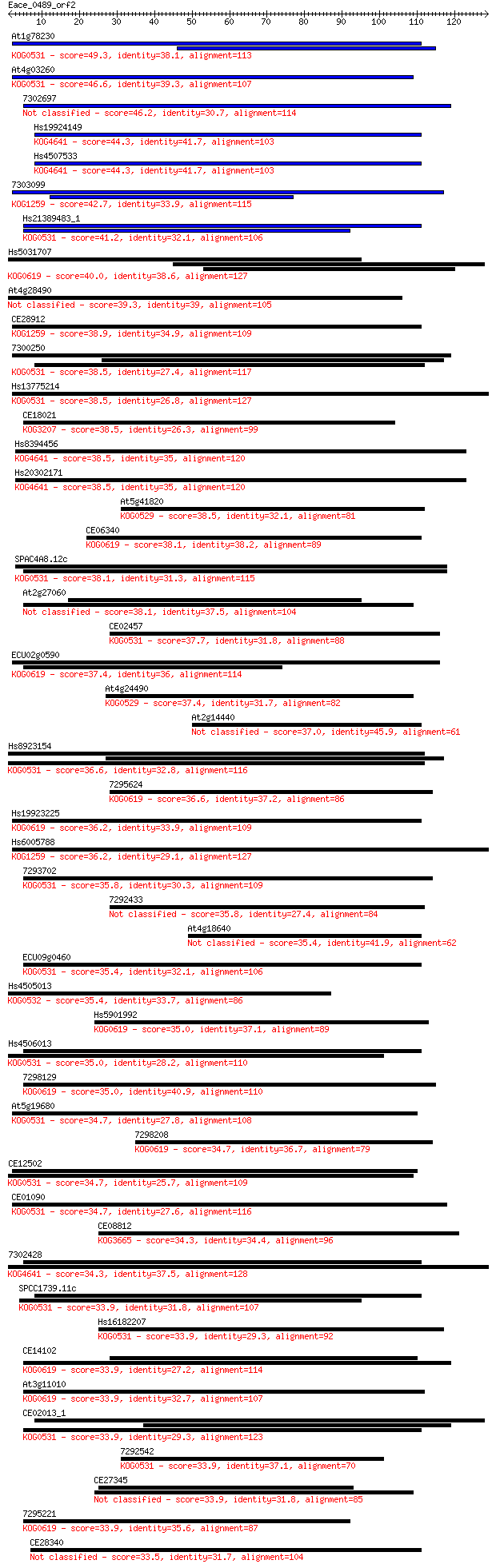
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0489_orf2
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
At1g78230 49.3 2e-06
At4g03260 46.6 1e-05
7302697 46.2 2e-05
Hs19924149 44.3 5e-05
Hs4507533 44.3 5e-05
7303099 42.7 2e-04
Hs21389483_1 41.2 4e-04
Hs5031707 40.0 0.001
At4g28490 39.3 0.002
CE28912 38.9 0.003
7300250 38.5 0.003
Hs13775214 38.5 0.003
CE18021 38.5 0.003
Hs8394456 38.5 0.003
Hs20302171 38.5 0.003
At5g41820 38.5 0.003
CE06340 38.1 0.004
SPAC4A8.12c 38.1 0.004
At2g27060 38.1 0.004
CE02457 37.7 0.005
ECU02g0590 37.4 0.006
At4g24490 37.4 0.007
At2g14440 37.0 0.010
Hs8923154 36.6 0.013
7295624 36.6 0.013
Hs19923225 36.2 0.017
Hs6005788 36.2 0.017
7293702 35.8 0.019
7292433 35.8 0.021
At4g18640 35.4 0.027
ECU09g0460 35.4 0.029
Hs4505013 35.4 0.029
Hs5901992 35.0 0.035
Hs4506013 35.0 0.035
7298129 35.0 0.039
At5g19680 34.7 0.040
7298208 34.7 0.046
CE12502 34.7 0.048
CE01090 34.7 0.051
CE08812 34.3 0.055
7302428 34.3 0.064
SPCC1739.11c 33.9 0.071
Hs16182207 33.9 0.072
CE14102 33.9 0.072
At3g11010 33.9 0.073
CE02013_1 33.9 0.077
7292542 33.9 0.077
CE27345 33.9 0.081
7295221 33.9 0.082
CE28340 33.5 0.094
> At1g78230
Length=581
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 42/110 (38%), Positives = 58/110 (52%), Gaps = 8/110 (7%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
F L +D++ N +V + SLP L AL S +I+ I RLR LD+S N+++
Sbjct 350 FTSLKSIDLSNNFIVQITPASLP-KGLHALNLSKNKISVIEGLRDLTRLRVLDLSYNRIS 408
Query 62 NV-INLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
+ LSN L ++EL LA NKIS +GL L L VLDL N +
Sbjct 409 RIGQGLSNCTL---IKELYLAGNKISNVEGLHRLL---KLIVLDLSFNKI 452
Score = 35.8 bits (81), Expect = 0.023, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 7/70 (10%)
Query 46 SHF-RLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLD 104
SHF L+ +D+S N ++ ++ L L LNL++NKIS +GL L+ L VLD
Sbjct 348 SHFTSLKSIDLSNN---FIVQITPASLPKGLHALNLSKNKISVIEGLRDLT---RLRVLD 401
Query 105 LRQNPLCNFG 114
L N + G
Sbjct 402 LSYNRISRIG 411
> At4g03260
Length=473
Score = 46.6 bits (109), Expect = 1e-05, Method: Composition-based stats.
Identities = 42/108 (38%), Positives = 59/108 (54%), Gaps = 8/108 (7%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
F L L+++GN +V + +LP L AL S I+ I RLR LD+S N++
Sbjct 214 FVGLRVLNLSGNAIVRITAGALP-RGLHALNLSKNSISVIEGLRELTRLRVLDLSYNRIL 272
Query 62 NVIN-LSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQN 108
+ + L++ +L+EL LA NKISE +GL L L VLDLR N
Sbjct 273 RLGHGLASCS---SLKELYLAGNKISEIEGLHRLL---KLTVLDLRFN 314
> 7302697
Length=261
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 64/115 (55%), Gaps = 4/115 (3%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
+T+L+++ S+ G + + L++L N + T+ F L++L++S N++++ +
Sbjct 17 ITELNLDNCRSTSIVGLTDEYTALESLSLINVGLTTLKGFPKLPNLKKLELSDNRISSGL 76
Query 65 N-LSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNVAE 118
N L+ +P L+ LNL+ NKI + + L PL F++L VLDL N N E
Sbjct 77 NYLTTSP---KLQYLNLSGNKIKDLETLKPLEEFKNLVVLDLFNNDATQVDNYRE 128
> Hs19924149
Length=839
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 43/129 (33%), Positives = 58/129 (44%), Gaps = 26/129 (20%)
Query 8 LDMNGNLLVSLDGHSL-PFPHLQALRASNCRINTITN--FGSHFRLRQLDISGNQL---- 60
LD++ N L L +S FP LQ L S C I TI + + S L L ++GN +
Sbjct 59 LDLSFNPLRHLGSYSFFSFPELQVLDLSRCEIQTIEDGAYQSLSHLSTLILTGNPIQSLA 118
Query 61 -----------------TNVINLSNTPLRF--TLRELNLARNKISEFKGLAPLSTFESLE 101
TN+ +L N P+ TL+ELN+A N I FK S +LE
Sbjct 119 LGAFSGLSSLQKLVAVETNLASLENFPIGHLKTLKELNVAHNLIQSFKLPEYFSNLTNLE 178
Query 102 VLDLRQNPL 110
LDL N +
Sbjct 179 HLDLSSNKI 187
> Hs4507533
Length=799
Score = 44.3 bits (103), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 43/129 (33%), Positives = 58/129 (44%), Gaps = 26/129 (20%)
Query 8 LDMNGNLLVSLDGHSL-PFPHLQALRASNCRINTITN--FGSHFRLRQLDISGNQL---- 60
LD++ N L L +S FP LQ L S C I TI + + S L L ++GN +
Sbjct 19 LDLSFNPLRHLGSYSFFSFPELQVLDLSRCEIQTIEDGAYQSLSHLSTLILTGNPIQSLA 78
Query 61 -----------------TNVINLSNTPLRF--TLRELNLARNKISEFKGLAPLSTFESLE 101
TN+ +L N P+ TL+ELN+A N I FK S +LE
Sbjct 79 LGAFSGLSSLQKLVAVETNLASLENFPIGHLKTLKELNVAHNLIQSFKLPEYFSNLTNLE 138
Query 102 VLDLRQNPL 110
LDL N +
Sbjct 139 HLDLSSNKI 147
> 7303099
Length=491
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 55/115 (47%), Gaps = 5/115 (4%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
++ +T+LD+ GNLL +DG P L+ L RI T+ N +L+ L +SGN +
Sbjct 311 WQEITELDLTGNLLTQIDGSVRTAPKLRRLILDQNRIRTVQNLAELPQLQLLSLSGNLIA 370
Query 62 NVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNV 116
++ T L L LA+NKI GL L SL LDL N + V
Sbjct 371 ECVDWHLT--MGNLVTLKLAQNKIKTLSGLRKLL---SLVNLDLSSNQIEELDEV 420
Score = 28.5 bits (62), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Query 12 GNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQ---LTNVINLSN 68
GNL+ L +L L+ + +I T++ L LD+S NQ L V +++N
Sbjct 366 GNLIAECVDWHLTMGNLVTLKLAQNKIKTLSGLRKLLSLVNLDLSSNQIEELDEVNHVAN 425
Query 69 TPLRFTLR 76
PL TLR
Sbjct 426 LPLLETLR 433
> Hs21389483_1
Length=440
Score = 41.2 bits (95), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 57/107 (53%), Gaps = 7/107 (6%)
Query 5 LTQLDMNGNLLVSLDG-HSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNV 63
L L + N + +++G + LP ++ L SN +I IT L+ LD+S NQ++++
Sbjct 239 LIHLSLANNKITTINGLNKLP---IKILCLSNNQIEMITGLEDLKALQNLDLSHNQISSL 295
Query 64 INLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
L N L L +NL NKI+E + + + L VL+L +NP+
Sbjct 296 QGLENHDL---LEVINLEDNKIAELREIEYIKNLPILRVLNLLENPI 339
Score = 35.0 bits (79), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 29/87 (33%), Positives = 43/87 (49%), Gaps = 4/87 (4%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
L +LD++ N + L S P+L L AS + T NF L++ D S NQ++ +
Sbjct 151 LQKLDLSANKIEDLSCVSC-MPYLLELNASQNNLTTFFNFKPPKNLKKADFSHNQISEIC 209
Query 65 NLSNTPLRFTLRELNLARNKISEFKGL 91
+LS L +L L N+I E GL
Sbjct 210 DLSAY---HALTKLILDGNEIEEISGL 233
> Hs5031707
Length=662
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 46/95 (48%), Gaps = 2/95 (2%)
Query 1 DFECLTQLDMNGNLLVSL-DGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQ 59
D L QLD++ N+L+ + DG P L L S + I++F S +LR LD+S N
Sbjct 171 DMPALEQLDLHSNVLMDIEDGAFEGLPRLTHLNLSRNSLTCISDF-SLQQLRVLDLSCNS 229
Query 60 LTNVINLSNTPLRFTLRELNLARNKISEFKGLAPL 94
+ S F L L+L NK+ F LA L
Sbjct 230 IEAFQTASQPQAEFQLTWLDLRENKLLHFPDLAAL 264
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 41/96 (42%), Gaps = 18/96 (18%)
Query 45 GSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLD 104
G L L + GN L ++ + P L+ LNLA N++S L + SLEVLD
Sbjct 488 GLEASLEVLALQGNGL--MVLQVDLPCFICLKRLNLAENRLSH---LPAWTQAVSLEVLD 542
Query 105 LRQN-----PLCNFGNVA--------EGFALVCCAN 127
LR N P G + +G L CC N
Sbjct 543 LRNNSFSLLPGSAMGGLETSLRRLYLQGNPLSCCGN 578
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 35/69 (50%), Gaps = 6/69 (8%)
Query 53 LDISGNQLTNVINLSNTPLRF--TLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
LD+SGNQL +++ +PL F LR L+L+ N+IS F LE L L N L
Sbjct 54 LDLSGNQLRSIL---ASPLGFYTALRHLDLSTNEIS-FLQPGAFQALTHLEHLSLAHNRL 109
Query 111 CNFGNVAEG 119
++ G
Sbjct 110 AMATALSAG 118
> At4g28490
Length=999
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 58/114 (50%), Gaps = 12/114 (10%)
Query 1 DFEC---LTQLDMNGNLLVSLDGHSLPF--PHLQALRAS--NCRINTITNFGSHFRLRQL 53
DF+ L LD++ NLLV SLPF P+L+ L S N ++FG +L L
Sbjct 109 DFDTCHNLISLDLSENLLVGSIPKSLPFNLPNLKFLEISGNNLSDTIPSSFGEFRKLESL 168
Query 54 DISGNQLTNVI--NLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDL 105
+++GN L+ I +L N TL+EL LA N S + + L L+VL L
Sbjct 169 NLAGNFLSGTIPASLGNV---TTLKELKLAYNLFSPSQIPSQLGNLTELQVLWL 219
> CE28912
Length=235
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 38/135 (28%), Positives = 60/135 (44%), Gaps = 31/135 (22%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHF----RLRQLDISG 57
++CL ++D++ N + D P ++ L N N+IT GS+ L +LD+S
Sbjct 26 WKCLEEVDLSFNEIKCFDESMKLLPEVRVL---NVSYNSITEIGSNLAFLSSLTELDLSN 82
Query 58 NQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESL----------------- 100
N LT I+ N L +++L L+ N I + GL L + E L
Sbjct 83 NTLTK-IDGWNEKLG-NIKKLILSENAIEDLSGLGKLYSLEYLDAKGNNIQNLDAVQGIG 140
Query 101 -----EVLDLRQNPL 110
E+L LR NP+
Sbjct 141 KLPCLEILLLRDNPI 155
> 7300250
Length=326
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/136 (23%), Positives = 59/136 (43%), Gaps = 23/136 (16%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
+ L +L++ N + ++ PHL+ L S R+ I N +L ++ N++T
Sbjct 82 LKTLIELELYDNQITKIENLD-DLPHLEVLDISFNRLTKIENLDKLVKLEKVYFVSNRIT 140
Query 62 NVINL-------------------SNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEV 102
+ NL N + LR+L L +NKI++ + L T +LE+
Sbjct 141 QIENLDMLTNLTMLELGDNKLKKIENIEMLVNLRQLFLGKNKIAKIEN---LDTLVNLEI 197
Query 103 LDLRQNPLCNFGNVAE 118
L L+ N + N+ +
Sbjct 198 LSLQANRIVKIENLEK 213
Score = 31.2 bits (69), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 44/91 (48%), Gaps = 6/91 (6%)
Query 26 PHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKI 85
P L ++ RI + NF R+ +L + N + + NLS+ TL EL L N+I
Sbjct 39 PDCYELDLNHRRIEKLENFEPLTRIERLFLRWNLIKKIENLSSLK---TLIELELYDNQI 95
Query 86 SEFKGLAPLSTFESLEVLDLRQNPLCNFGNV 116
++ + L L LEVLD+ N L N+
Sbjct 96 TKIENLDDLP---HLEVLDISFNRLTKIENL 123
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 49/104 (47%), Gaps = 4/104 (3%)
Query 8 LDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLS 67
L + N +V ++ +L+ L S + TI N + +L LD++ N+L + +
Sbjct 198 LSLQANRIVKIENLE-KLANLRELYVSENGVETIENLSENTKLETLDLAKNRLKGI---A 253
Query 68 NTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLC 111
N L EL L N + ++K + L ++L+ + L NPL
Sbjct 254 NLEKLELLEELWLNHNGVDDWKDIELLKVNKALQTIYLEYNPLA 297
> Hs13775214
Length=225
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 34/127 (26%), Positives = 57/127 (44%), Gaps = 8/127 (6%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
FE L +L ++ N++ ++G HL L S I TI + L L + N+++
Sbjct 64 FENLRKLQLDNNIIEKIEGLE-NLAHLVWLDLSFNNIETIEGLDTLVNLEDLSLFNNRIS 122
Query 62 NVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNVAEGFA 121
+ +L L+ L+L N+I + L F+ L L L +NP+ AE +
Sbjct 123 KIDSLDAL---VKLQVLSLGNNRIDNMMNIIYLRRFKCLRTLSLSRNPISE----AEDYK 175
Query 122 LVCCANL 128
+ CA L
Sbjct 176 MFICAYL 182
> CE18021
Length=493
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 42/99 (42%), Gaps = 22/99 (22%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
LT LD+ N +LD FP+L L +NC I ++ F + +L+
Sbjct 251 LTSLDLEDNPFKTLDSIHGTFPNLTQLSVANCGITSLNGFDGSIKFPKLEY--------- 301
Query 65 NLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVL 103
LN+ RN I E+K + + + +SL+ L
Sbjct 302 -------------LNIKRNSIVEWKSVNSIRSLKSLKRL 327
> Hs8394456
Length=1032
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 42/130 (32%), Positives = 63/130 (48%), Gaps = 17/130 (13%)
Query 3 ECLTQLDMNGNLLVSLDGHSLPF-PHLQALRASNCRINTITN--FGSHFRLRQLDISGNQ 59
+ L L + N L SL F P L+ L + ++ +TN + RLR+LD+S N
Sbjct 653 KSLQVLRLRDNYLAFFKWWSLHFLPKLEVLDLAGNQLKALTNGSLPAGTRLRRLDVSCNS 712
Query 60 LTNVINLSNTPLRFT----LRELNLARN--KISEFKGLAPLSTFESLEVLDLRQNPL-CN 112
++ V P F+ LRELNL+ N K + PL++ +L++LD+ NPL C
Sbjct 713 ISFV-----APGFFSKAKELRELNLSANALKTVDHSWFGPLAS--ALQILDVSANPLHCA 765
Query 113 FGNVAEGFAL 122
G F L
Sbjct 766 CGAAFMDFLL 775
> Hs20302171
Length=975
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 42/130 (32%), Positives = 63/130 (48%), Gaps = 17/130 (13%)
Query 3 ECLTQLDMNGNLLVSLDGHSLPF-PHLQALRASNCRINTITN--FGSHFRLRQLDISGNQ 59
+ L L + N L SL F P L+ L + ++ +TN + RLR+LD+S N
Sbjct 596 KSLQVLRLRDNYLAFFKWWSLHFLPKLEVLDLAGNQLKALTNGSLPAGTRLRRLDVSCNS 655
Query 60 LTNVINLSNTPLRFT----LRELNLARN--KISEFKGLAPLSTFESLEVLDLRQNPL-CN 112
++ V P F+ LRELNL+ N K + PL++ +L++LD+ NPL C
Sbjct 656 ISFV-----APGFFSKAKELRELNLSANALKTVDHSWFGPLAS--ALQILDVSANPLHCA 708
Query 113 FGNVAEGFAL 122
G F L
Sbjct 709 CGAAFMDFLL 718
> At5g41820
Length=687
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 3/81 (3%)
Query 31 LRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKG 90
LR +N ++ I ++ LD+S N+L + L L L LNL+ N+I F
Sbjct 529 LRLNNLTLSRIAAVEKLLFVQMLDLSHNELHSAEGLEAMQL---LCCLNLSHNRIRSFSA 585
Query 91 LAPLSTFESLEVLDLRQNPLC 111
L L + L VLD+ N +C
Sbjct 586 LDSLRHLKQLRVLDVSHNHIC 606
> CE06340
Length=603
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 48/91 (52%), Gaps = 4/91 (4%)
Query 22 SLPFPHLQAL--RASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELN 79
S+ P L+ L ++N T++NFG +LR LD+S N L + T LR LR L+
Sbjct 107 SIRLPSLEVLDLHSNNIEHATMSNFGGMPKLRVLDLSSNHLNILPTGVFTYLR-ALRSLS 165
Query 80 LARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
L+ N IS+ L SL VL L +NP+
Sbjct 166 LSNNTISDLST-NLLRGLNSLRVLRLDRNPI 195
> SPAC4A8.12c
Length=332
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 36/136 (26%), Positives = 62/136 (45%), Gaps = 27/136 (19%)
Query 3 ECLTQLDMNGNLLVSLDG---------HSLPFPHLQALRASNC------------RINTI 41
E LT+LD+ NL+V ++ L F +++ +R N RI I
Sbjct 82 ETLTELDLYDNLIVRIENLDNVKNLTYLDLSFNNIKTIRNINHLKGLENLFFVQNRIRRI 141
Query 42 TNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLE 101
N RL L++ GN++ + NL L +L + +NKI++F+ L + L
Sbjct 142 ENLEGLDRLTNLELGGNKIRVIENLDTL---VNLEKLWVGKNKITKFENFEKL---QKLS 195
Query 102 VLDLRQNPLCNFGNVA 117
+L ++ N + F N+A
Sbjct 196 LLSIQSNRITQFENLA 211
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 56/113 (49%), Gaps = 6/113 (5%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
LT L++ GN + ++ +L+ L +I NF +L L I N++T
Sbjct 150 LTNLELGGNKIRVIENLD-TLVNLEKLWVGKNKITKFENFEKLQKLSLLSIQSNRITQFE 208
Query 65 NLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNVA 117
NL+ L LREL ++ N ++ F G+ L E+LE+LD+ N + + +A
Sbjct 209 NLA--CLSHCLRELYVSHNGLTSFSGIEVL---ENLEILDVSNNMIKHLSYLA 256
> At2g27060
Length=1007
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 45/80 (56%), Gaps = 6/80 (7%)
Query 17 SLDGHSLPFPHLQALRASNCRINTITNF--GSHFRLRQLDISGNQLTNVINLSNTPLRFT 74
+L G + F L +L+A+N + + F G++ L+++D+S NQL+ VI SN +
Sbjct 360 TLPGQTSQFLRLTSLKAANNSLQGVLPFILGTYPELKEIDLSHNQLSGVIP-SNLFISAK 418
Query 75 LRELNLARNKISEFKGLAPL 94
L ELNL+ N F G PL
Sbjct 419 LTELNLSNNN---FSGSLPL 435
Score = 30.8 bits (68), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 16/113 (14%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALR-------ASNCRINTITNFGSHFRLRQLDISG 57
+T +D+NG L+ S FP + LR A+N T++N GS L+ LD+SG
Sbjct 57 VTSIDLNGFGLLG----SFSFPVIVGLRMLQNLSIANNQFSGTLSNIGSLTSLKYLDVSG 112
Query 58 NQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAP--LSTFESLEVLDLRQN 108
N + LR L +NL+ N + G+ P + L+ LDL+ N
Sbjct 113 NLFHGALPSGIENLR-NLEFVNLSGN--NNLGGVIPSGFGSLAKLKYLDLQGN 162
> CE02457
Length=349
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 42/88 (47%), Gaps = 3/88 (3%)
Query 28 LQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISE 87
L L + +I + N S L LD+S N++T + NL L+ L NKI++
Sbjct 78 LTHLEFYDNQITKVENLDSLVNLESLDLSFNRITKIENLEKLT---KLKTLFFVHNKITK 134
Query 88 FKGLAPLSTFESLEVLDLRQNPLCNFGN 115
+GL L+ E LE+ D R + N N
Sbjct 135 IEGLDTLTELEYLELGDNRIAKIENLDN 162
> ECU02g0590
Length=785
Score = 37.4 bits (85), Expect = 0.006, Method: Composition-based stats.
Identities = 41/114 (35%), Positives = 54/114 (47%), Gaps = 17/114 (14%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
FE L L MN + L SL L+ L+A N I I S L LD+SGN
Sbjct 272 FESLVFLSMNDCGVYCL---SLTARKLRVLKARNNFIEEIPLLPS---LEYLDVSGNL-- 323
Query 62 NVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGN 115
V +S+ +RF LN ++N+I F LS++ L LDL NPL + N
Sbjct 324 -VHKISSKGVRF----LNASKNQIMTFD----LSSWNRLVHLDLSYNPLVSLEN 368
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 24/69 (34%), Positives = 35/69 (50%), Gaps = 6/69 (8%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
L LD++GNL+ H + ++ L AS +I T + S RL LD+S N L ++
Sbjct 314 LEYLDVSGNLV-----HKISSKGVRFLNASKNQIMTF-DLSSWNRLVHLDLSYNPLVSLE 367
Query 65 NLSNTPLRF 73
N LRF
Sbjct 368 NSDARGLRF 376
> At4g24490
Length=683
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 41/82 (50%), Gaps = 3/82 (3%)
Query 27 HLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKIS 86
+L LR +N ++ I + ++ LD+S N+L + L L L LNL+ N+I
Sbjct 517 NLVCLRLNNLSLSRIASVEKLLFVQMLDLSHNELHSTEGLEAMQL---LSCLNLSHNRIR 573
Query 87 EFKGLAPLSTFESLEVLDLRQN 108
F L L + L+VLD+ N
Sbjct 574 SFSALDSLRHVKQLKVLDVSHN 595
> At2g14440
Length=886
Score = 37.0 bits (84), Expect = 0.010, Method: Composition-based stats.
Identities = 28/63 (44%), Positives = 37/63 (58%), Gaps = 6/63 (9%)
Query 50 LRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAP--LSTFESLEVLDLRQ 107
LR+LD+S N LT VI S L LREL+L+ N ++ G P L+T + L V+ LR
Sbjct 438 LRELDLSNNNLTGVIPPSLQNLTM-LRELDLSNNNLT---GEVPEFLATIKPLLVIHLRG 493
Query 108 NPL 110
N L
Sbjct 494 NNL 496
> Hs8923154
Length=685
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 37/111 (33%), Positives = 55/111 (49%), Gaps = 7/111 (6%)
Query 1 DFECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQL 60
+ + L LD+ N + + G S L+ L RI I+N + L LD+ GNQ+
Sbjct 132 NLQKLISLDLYDNQIEEISGLS-TLRCLRVLLLGKNRIKKISNLENLKSLDVLDLHGNQI 190
Query 61 TNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLC 111
T + N+++ LR LNLARN +S L L +SL L+LR N +
Sbjct 191 TKIENINHL---CELRVLNLARNFLSHVDNLNGL---DSLTELNLRHNQIT 235
Score = 34.3 bits (77), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 46/90 (51%), Gaps = 6/90 (6%)
Query 27 HLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKIS 86
HL+ L + I I N + +L LD+ NQ+ + LS LR LR L L +N+I
Sbjct 113 HLRLLNFQHNFITRIQNISNLQKLISLDLYDNQIEEISGLST--LR-CLRVLLLGKNRI- 168
Query 87 EFKGLAPLSTFESLEVLDLRQNPLCNFGNV 116
K ++ L +SL+VLDL N + N+
Sbjct 169 --KKISNLENLKSLDVLDLHGNQITKIENI 196
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 54/118 (45%), Gaps = 18/118 (15%)
Query 1 DFECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHF-------RLRQL 53
+ + L LD++GN + ++ HL LR +N NF SH L +L
Sbjct 176 NLKSLDVLDLHGNQITKIEN----INHLCELRV----LNLARNFLSHVDNLNGLDSLTEL 227
Query 54 DISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLC 111
++ NQ+T V ++ N P L+ L L+ N IS F ++ L+ SL NP+
Sbjct 228 NLRHNQITFVRDVDNLP---CLQHLFLSFNNISSFDSVSCLADSSSLSDTTFDGNPIA 282
> 7295624
Length=1470
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 32/88 (36%), Positives = 47/88 (53%), Gaps = 4/88 (4%)
Query 28 LQALRASNCRINTITN--FGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKI 85
LQ L SNC + I + F + L +LD+ N+LT IN + +EL L RN++
Sbjct 598 LQQLSMSNCSLGQIPDLLFAKNTNLVRLDLCDNRLTQ-INRNIFSGLNVFKELRLCRNEL 656
Query 86 SEFKGLAPLSTFESLEVLDLRQNPLCNF 113
S+F +A L +LE LDL +N L +
Sbjct 657 SDFPHIA-LYNLSTLESLDLARNQLASI 683
> Hs19923225
Length=440
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 37/111 (33%), Positives = 56/111 (50%), Gaps = 10/111 (9%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTI--TNFGSHFRLRQLDISGNQ 59
+ L LD++ N L SL H LP L + A+N I + ++ + L+ LD+S N
Sbjct 77 YTNLRTLDISNNRLESLPAH-LPR-SLWNMSAANNNIKLLDKSDTAYQWNLKYLDVSKNM 134
Query 60 LTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
L V+ + NT LR +L LNL+ NK+ P + L ++DL N L
Sbjct 135 LEKVVLIKNT-LR-SLEVLNLSSNKL----WTVPTNMPSKLHIVDLSNNSL 179
> Hs6005788
Length=1504
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 62/127 (48%), Gaps = 5/127 (3%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
++ LT LD++ N + +D P ++ L S+ + + N + L LD+S N+L+
Sbjct 286 WQALTTLDLSHNSISEIDESVKLIPKIEFLDLSHNGLLVVDNLQHLYNLVHLDLSYNKLS 345
Query 62 NVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNVAEGFA 121
++ L +T L ++ LNLA N + GL L SL LDLR N + V +
Sbjct 346 SLEGL-HTKLG-NIKTLNLAGNLLESLSGLHKLY---SLVNLDLRDNRIEQMEEVRSIGS 400
Query 122 LVCCANL 128
L C ++
Sbjct 401 LPCLEHV 407
> 7293702
Length=392
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 49/109 (44%), Gaps = 3/109 (2%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
L LD++G++L SL L L SNC +N+ +R L GN + V
Sbjct 136 LQSLDLSGSVLSSLRDLGYGLLQLTRLDISNCGLNSFDGTSGLPAIRVLIADGNMIQRVD 195
Query 65 NLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNF 113
L+ LR L N+ISE L+ L L+ ++L+ NP+C
Sbjct 196 PLAEL---VHLRVLKARNNRISELGLLSFLGMCPQLQEVELQGNPVCRL 241
> 7292433
Length=454
Score = 35.8 bits (81), Expect = 0.021, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 40/84 (47%), Gaps = 0/84 (0%)
Query 28 LQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISE 87
L ++ NC + +++ R+R +++ + + LS L+EL L +N IS+
Sbjct 18 LSLIKKLNCWGSDLSDVSIIKRMRGVEVLALSVNKISTLSTFEDCTKLQELYLRKNSISD 77
Query 88 FKGLAPLSTFESLEVLDLRQNPLC 111
+A L SL L L +NP C
Sbjct 78 INEIAYLQNLPSLRNLWLEENPCC 101
> At4g18640
Length=685
Score = 35.4 bits (80), Expect = 0.027, Method: Composition-based stats.
Identities = 26/64 (40%), Positives = 36/64 (56%), Gaps = 6/64 (9%)
Query 49 RLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAP--LSTFESLEVLDLR 106
+++ LD+SG L + + L LR L L+RN F G P +FE+LEVLDLR
Sbjct 70 KVQILDLSGYSLEGTLAPELSQLS-DLRSLILSRN---HFSGGIPKEYGSFENLEVLDLR 125
Query 107 QNPL 110
+N L
Sbjct 126 ENDL 129
> ECU09g0460
Length=218
Score = 35.4 bits (80), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 55/106 (51%), Gaps = 10/106 (9%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
L LD++ NL+ + S+P +L+ L + I TI R+++LD++ N + +
Sbjct 65 LKVLDLSYNLITDI---SIPPMNLEELYLISNDIATIHGLNLP-RIKKLDMAVNDICKIE 120
Query 65 NLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
NL TL EL L N+I +GL + SL++LDL+ N L
Sbjct 121 NLEKCT---TLEELYLGSNQIGAVEGLEEMR---SLKILDLQNNKL 160
> Hs4505013
Length=832
Score = 35.4 bits (80), Expect = 0.029, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 49/91 (53%), Gaps = 12/91 (13%)
Query 1 DFECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTIT-NFGSHFRLRQLDISGNQ 59
+ LT L+++ N L L + P L+ L SN ++ + + G+ LRQLD+S N+
Sbjct 112 NLTALTYLNLSRNQLSLLPPYICQLP-LRVLIVSNNKLGALPPDIGTLGSLRQLDVSSNE 170
Query 60 L----TNVINLSNTPLRFTLRELNLARNKIS 86
L + + LS +LR+LN+ RN++S
Sbjct 171 LQSLPSELCGLS------SLRDLNVRRNQLS 195
> Hs5901992
Length=352
Score = 35.0 bits (79), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 46/96 (47%), Gaps = 12/96 (12%)
Query 24 PFPHLQALRASNCRINTITNFG-------SHFRLRQLDISGNQLTNVINLSNTPLRFTLR 76
PF + LR N N ITN+G +L L + N+L V +PL +L
Sbjct 90 PFENATQLRWINLNKNKITNYGIEKGALSQLKKLLFLFLEDNELEEV----PSPLPRSLE 145
Query 77 ELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCN 112
+L LARNK+S S E+L +LDL+ N L +
Sbjct 146 QLQLARNKVSRIPQ-GTFSNLENLTLLDLQNNKLVD 180
> Hs4506013
Length=360
Score = 35.0 bits (79), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 51/106 (48%), Gaps = 4/106 (3%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
LT L M N L ++G +L+ L S+ I I ++ +L LDI+ N++ +
Sbjct 232 LTVLSMQSNRLTKIEGLQ-NLVNLRELYLSHNGIEVIEGLENNNKLTMLDIASNRIKKIE 290
Query 65 NLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
N+S+ L+E + N + + L L SLE + L +NPL
Sbjct 291 NISHLT---ELQEFWMNDNLLESWSDLDELKGARSLETVYLERNPL 333
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 46/100 (46%), Gaps = 4/100 (4%)
Query 1 DFECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQL 60
+ + L +LD+ N + ++ L+ L S + I RL++L + N++
Sbjct 118 ELQSLRELDLYDNQIKKIENLE-ALTELEILDISFNLLRNIEGVDKLTRLKKLFLVNNKI 176
Query 61 TNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESL 100
+ + NLSN L+ L L N+I + + L+ ESL
Sbjct 177 SKIENLSNL---HQLQMLELGSNRIRAIENIDTLTNLESL 213
> 7298129
Length=573
Score = 35.0 bits (79), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 45/137 (32%), Positives = 60/137 (43%), Gaps = 32/137 (23%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRA----SNCRINTITNFGSHFR-LRQLDISGNQ 59
LT L +NGN LVSLDG L HLQ LR N TN + R L L + NQ
Sbjct 229 LTSLQLNGNALVSLDGDCL--GHLQKLRTLRLEGNLFYRIPTNALAGLRTLEALLLKRNQ 286
Query 60 LTNVINLSNTPLR--FTLRELNLARNKISEF-KGLAPLSTFE------------------ 98
++ +S L+ L+ L L N IS +GL+ LS +
Sbjct 287 ---IMKISAGALKNLTALKVLELDDNLISSLPEGLSKLSQLQELSITSNRLRWINDTELP 343
Query 99 -SLEVLDLRQNPLCNFG 114
S+++LD+R NPL
Sbjct 344 RSMQMLDMRANPLSTIS 360
> At5g19680
Length=328
Score = 34.7 bits (78), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 52/110 (47%), Gaps = 6/110 (5%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
+C+ ++ + N L S+ G L+ L S+ I+ + + LR LD+S N+LT
Sbjct 196 LKCIKKISLQSNRLTSMKGFE-ECVALEELYLSHNGISKMEGLSALVNLRVLDVSNNKLT 254
Query 62 NVINLSNTPLRFTLRELNLARNKISEFKGL--APLSTFESLEVLDLRQNP 109
+V ++ N L +L L N+I + + A + E L + L NP
Sbjct 255 SVDDIQNLT---KLEDLWLNDNQIESLEAITEAVTGSKEKLTTIYLENNP 301
> 7298208
Length=1043
Score = 34.7 bits (78), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 42/83 (50%), Gaps = 7/83 (8%)
Query 35 NCRINTITNF----GSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKG 90
N N I +F S+ + QLDIS N +T + +N L TLR LNLA N++ +
Sbjct 856 NVSHNHIASFYDELSSYMYIYQLDISHNHVTKSDSFTN--LANTLRFLNLAHNQLGSLQS 913
Query 91 LAPLSTFESLEVLDLRQNPLCNF 113
A E LE+L++ N L +
Sbjct 914 HA-FGDLEFLEILNVAHNNLTSL 935
> CE12502
Length=630
Score = 34.7 bits (78), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 54/108 (50%), Gaps = 2/108 (1%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
FE L LD++ NL+ SL +L+ L S +N I + L+ + ++ N+++
Sbjct 141 FENLKILDLSNNLIEPPVSFSLK--NLEILNLSGNFLNEIPDLSKCVALQTISLADNKIS 198
Query 62 NVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNP 109
++ ++ L+ L+++ N I + + LSTF+ LE + NP
Sbjct 199 DLTTITKLICPTNLKNLDISSNSIEDLSQFSVLSTFKKLEEFVVAGNP 246
Score = 28.9 bits (63), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 45/109 (41%), Gaps = 32/109 (29%)
Query 1 DFECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINT-ITNFGSHFRLRQLDISGNQ 59
++ + L ++GN L FP Q + A +C+IN I +F
Sbjct 75 EYSEVKHLKISGNCFQKFTYIKL-FPKCQIIDARDCQINKFIADFN-------------- 119
Query 60 LTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQN 108
+ L EL+LARN++ E L FE+L++LDL N
Sbjct 120 -------------YNLLELHLARNQLKETNQLG---RFENLKILDLSNN 152
> CE01090
Length=326
Score = 34.7 bits (78), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 32/116 (27%), Positives = 52/116 (44%), Gaps = 3/116 (2%)
Query 2 FECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLT 61
F + +L M NLLVS+ L +L ++ I++ S L LD+S N++
Sbjct 57 FPKIEELRMRNNLLVSISPTISSLVTLTSLDLYENQLTEISHLESLVNLVSLDLSYNRIR 116
Query 62 NVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNVA 117
+ L L L L NKI + + L L+ + LE+ D R + N G++
Sbjct 117 QINGLDKLT---KLETLYLVSNKIEKIENLEALTQLKLLELGDNRIKKIENIGHLV 169
> CE08812
Length=586
Score = 34.3 bits (77), Expect = 0.055, Method: Composition-based stats.
Identities = 33/101 (32%), Positives = 45/101 (44%), Gaps = 8/101 (7%)
Query 25 FPHLQALRASNCRINT---ITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLA 81
P L L S+ IN FG+ LR LDISG TN++N++ L L L
Sbjct 147 LPSLTHLTMSDVHINEDDFYQLFGNFSNLRSLDISG---TNILNIAGISRLKQLEVLALR 203
Query 82 RNKISEFKGLAPLSTFESLEVLDLRQN--PLCNFGNVAEGF 120
I+ + L L + L VLD+ Q L + N+ E F
Sbjct 204 DFSIATYTELKDLFNLKHLRVLDVSQTRFNLHSRNNIVEQF 244
> 7302428
Length=1446
Score = 34.3 bits (77), Expect = 0.064, Method: Compositional matrix adjust.
Identities = 38/119 (31%), Positives = 52/119 (43%), Gaps = 14/119 (11%)
Query 5 LTQLDMNGNLLVSL-DGHSLPFPHLQALRASNCRINTITNFG------------SHFRLR 51
LT LD+ N L L G P +LQ L + RI T G + L+
Sbjct 191 LTDLDLGDNNLRQLPSGFLCPVGNLQVLNLTRNRIRTAEQMGFADMNCGAGSGSAGSELQ 250
Query 52 QLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
LD S N+L ++ L+ LNLA N +SE G A L+ SL +++L N L
Sbjct 251 VLDASHNELRSISESWGISRLRRLQHLNLAYNNLSELSGEA-LAGLASLRIVNLSNNHL 308
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 37/131 (28%), Positives = 61/131 (46%), Gaps = 14/131 (10%)
Query 1 DFECLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHF---RLRQLDISG 57
+FE L + ++ N L ++G F L +L N N + F F L+ LDI G
Sbjct 560 NFE-LEAIRLDRNFLADING---VFATLVSLLWLNLSENHLVWFDYAFIPSNLKWLDIHG 615
Query 58 NQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNVA 117
N + + N ++ L+ + N+I+E + P+S ++E+L + N + GNV
Sbjct 616 NYIEALGNYYKLQEEIRVKTLDASHNRITE---IGPMSIPNTIELLFINNNLI---GNVQ 669
Query 118 EGFALVCCANL 128
A V ANL
Sbjct 670 PN-AFVDKANL 679
> SPCC1739.11c
Length=1045
Score = 33.9 bits (76), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 57/106 (53%), Gaps = 7/106 (6%)
Query 8 LDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNF--GSHFRLRQLDISGNQLTNVIN 65
L ++ N V+LD + F ++ L +N ++ + + S LR LD+S N ++++ +
Sbjct 850 LYLSNNTFVTLDCKHM-FLGVRYLELANVQLKEVPKYIATSMPNLRVLDLSHNYISDIES 908
Query 66 LSNTPLRFTLRELNLARNKISEFKGLAP-LSTFESLEVLDLRQNPL 110
L PL+ R L L N+I + + L L+ + L VLDLR NPL
Sbjct 909 L--KPLQMIHR-LYLVGNRIKKMRNLCDILANLKQLNVLDLRMNPL 951
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 47/91 (51%), Gaps = 6/91 (6%)
Query 4 CLTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNV 63
+ +L + GN + L G + ++ L A R++++T+F + L+ LDIS NQL ++
Sbjct 627 SIEELTLEGNEIAYLTGCPVT---IRDLNAVENRLSSLTSFSNLLNLQYLDISYNQLEDL 683
Query 64 INLSNTPLRFTLRELNLARNKISEFKGLAPL 94
LS+ LREL + N + G+ L
Sbjct 684 TGLSSL---IHLRELKVDSNHLWSLDGIQHL 711
> Hs16182207
Length=294
Score = 33.9 bits (76), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 49/93 (52%), Gaps = 7/93 (7%)
Query 25 FPHLQALRASNCRINTITNFGSHF-RLRQLDISGNQLTNVINLSNTPLRFTLRELNLARN 83
P+L L+ + + ++ + G+ L+ L ++ L ++ +++ P L+EL + N
Sbjct 92 LPNLDQLKLNGSHLGSLRDLGTSLGHLQVLWLARCGLADLDGIASLP---ALKELYASYN 148
Query 84 KISEFKGLAPLSTFESLEVLDLRQNPLCNFGNV 116
IS+ L+PL E LEVLDL N + + G V
Sbjct 149 NISD---LSPLCLLEQLEVLDLEGNSVEDLGQV 178
> CE14102
Length=653
Score = 33.9 bits (76), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 21/84 (25%), Positives = 44/84 (52%), Gaps = 4/84 (4%)
Query 28 LQALRASNCRINTIT--NFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKI 85
L+ L + C +I+ + +L +LD+S ++N++ + + +L++LNL +NK+
Sbjct 376 LEKLSLAECGFTSISADQLKDYPKLEELDLSKCNISNIVENTFENQKDSLKKLNLQKNKL 435
Query 86 SEFKGLAPLSTFESLEVLDLRQNP 109
L + ++E LD+ NP
Sbjct 436 KSLPNL--IKNLPAIESLDVSSNP 457
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 36/143 (25%), Positives = 58/143 (40%), Gaps = 38/143 (26%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFR---------LRQLDI 55
L +LD+ NL+ ++ ++ F L++L R++ NF S L +LD+
Sbjct 282 LKELDLRVNLIENITAYA--FDGLESL----TRLSLAGNFISKLEPDVFFGLSSLEELDL 335
Query 56 SGNQLTNVINLSNTPLRFTLRELNLARNKISEFK--GLA------------------PLS 95
N++ + PL L+ ++L N ISE GL L
Sbjct 336 GWNEIKTIPTDVFKPLTDKLKTISLRNNPISELPSTGLGMLEKLSLAECGFTSISADQLK 395
Query 96 TFESLEVLDLRQNPLCNFGNVAE 118
+ LE LDL + CN N+ E
Sbjct 396 DYPKLEELDLSK---CNISNIVE 415
> At3g11010
Length=957
Score = 33.9 bits (76), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 48/109 (44%), Gaps = 5/109 (4%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRI-NTITNFGSHFR-LRQLDISGNQLTN 62
L L+++ N + + P P + L SN I +F R L LD+S N +
Sbjct 537 LFYLNLSNNTFIGFQRPTKPEPSMAYLLGSNNNFTGKIPSFICELRSLYTLDLSDNNFSG 596
Query 63 VINLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLC 111
I L+ L ELNL +N +S G P FESL LD+ N L
Sbjct 597 SIPRCMENLKSNLSELNLRQNNLS---GGFPEHIFESLRSLDVGHNQLV 642
> CE02013_1
Length=296
Score = 33.9 bits (76), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 53/120 (44%), Gaps = 4/120 (3%)
Query 8 LDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLS 67
L + NLL +D H L L ++ +I + N + L LD+S N++T + LS
Sbjct 15 LSLRWNLLKKID-HFQCLTSLTRLNLNDNQIEKLENLETLVNLVFLDVSYNRITKIEGLS 73
Query 68 NTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPLCNFGNVAEGFALVCCAN 127
L EL+L NKI +GL + + LE D R + N ++ L AN
Sbjct 74 EL---INLEELHLVHNKIITIEGLETNTAMKYLEFGDNRIQKMENLSHLVNLERLFLGAN 130
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 6/82 (7%)
Query 37 RINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKISEFKGLAPLST 96
+I I +L++L + GN L + L L+ ++LA+N I + GL+ L+
Sbjct 131 QIRKIEGLDGMAQLKELSLPGNALQIIEGLDTLS---GLKSISLAQNGIRKIDGLSGLTN 187
Query 97 FESLEVLDLRQNPLCNFGNVAE 118
+S LDL N + NV +
Sbjct 188 LKS---LDLNDNIIEKLENVEQ 206
Score = 27.3 bits (59), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 48/106 (45%), Gaps = 4/106 (3%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVI 64
L +L + GN L ++G L+++ + I I L+ LD++ N +
Sbjct 144 LKELSLPGNALQIIEGLD-TLSGLKSISLAQNGIRKIDGLSGLTNLKSLDLNDNIIEK-- 200
Query 65 NLSNTPLRFTLRELNLARNKISEFKGLAPLSTFESLEVLDLRQNPL 110
L N + L + +NK++ ++ + L E+L VL + NPL
Sbjct 201 -LENVEQFKGISSLMIRKNKLNCWQDVRQLKKLENLTVLTMEMNPL 245
> 7292542
Length=1301
Score = 33.9 bits (76), Expect = 0.077, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 37/74 (50%), Gaps = 5/74 (6%)
Query 31 LRASNCRINTITNFGSHF----RLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNKIS 86
LR + N +T+ GS +L+ L+++GNQ+ + LR LRELNL RNK+
Sbjct 517 LRVLDLHGNKLTSLGSRINCLQQLKSLNLAGNQIRQINQQDFLGLR-CLRELNLKRNKLR 575
Query 87 EFKGLAPLSTFESL 100
G L E L
Sbjct 576 RINGFQHLVALERL 589
> CE27345
Length=773
Score = 33.9 bits (76), Expect = 0.081, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 40/68 (58%), Gaps = 6/68 (8%)
Query 25 FPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRFTLRELNLARNK 84
FP +Q+L S S+ +L+ LD+S N++++ + + NT LR LN++RN+
Sbjct 145 FPKIQSLSLSKNYFEKFECDTSNTKLKILDLSQNRISH-LEVPNT-----LRVLNVSRNR 198
Query 85 ISEFKGLA 92
++ F+ ++
Sbjct 199 LTSFENIS 206
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 21/86 (24%), Positives = 43/86 (50%), Gaps = 4/86 (4%)
Query 24 PFPHLQALRASNCRINTITNFGSHFRLRQLDISGNQLTNVINLSNTPLRF-TLRELNLAR 82
PFP L+ + + + ++ H L+ L I ++LT+ + F ++ L+L++
Sbjct 96 PFPQLEHIILRDGDLESLNGTVIHPTLKVLSIENSELTSSSEVCRLLSIFPKIQSLSLSK 155
Query 83 NKISEFKGLAPLSTFESLEVLDLRQN 108
N +F+ ++ L++LDL QN
Sbjct 156 NYFEKFEC---DTSNTKLKILDLSQN 178
> 7295221
Length=724
Score = 33.9 bits (76), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 49/93 (52%), Gaps = 7/93 (7%)
Query 5 LTQLDMNGNLLVSLDGHSLPFPHLQALRASNCRIN--TITNFGSHFRLRQLDISGNQLTN 62
L LD++ N + SL+G +L+ L + R+ + +F RL LD++GNQL
Sbjct 276 LVNLDLDNNRIDSLNGALAGLGNLRILNLAGNRLEHLQVGDFDGMIRLDILDLTGNQLAE 335
Query 63 VINLSNTPLRFTLRELNLARNKIS----EFKGL 91
+ L T L +L+ L +A N I+ +FKGL
Sbjct 336 LKPLEMT-LLPSLKILKVAYNNITKLEQDFKGL 367
> CE28340
Length=444
Score = 33.5 bits (75), Expect = 0.094, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 53/110 (48%), Gaps = 15/110 (13%)
Query 7 QLDMNGNLLVSLDGHSLPFPHLQALRASNCRINTITNFGSHF----RLRQLDISGNQLTN 62
+LD++ + + + P L L S+ N IT F +L +LD QL +
Sbjct 19 ELDLSASGIQEFPNAIVQLPRLTKLDLSS---NAITFLPESFCKMTKLIRLDFGSCQLHH 75
Query 63 VINLSNTPLRFTLRELNLARNKISEFKGLAPLS--TFESLEVLDLRQNPL 110
+ + L +L+ LNL N+I + PLS +SL+ LDL++NPL
Sbjct 76 LPD--GIGLLTSLQHLNLYNNQIEDL----PLSFANLKSLKWLDLKKNPL 119
Lambda K H
0.324 0.139 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40