bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eace_0497_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
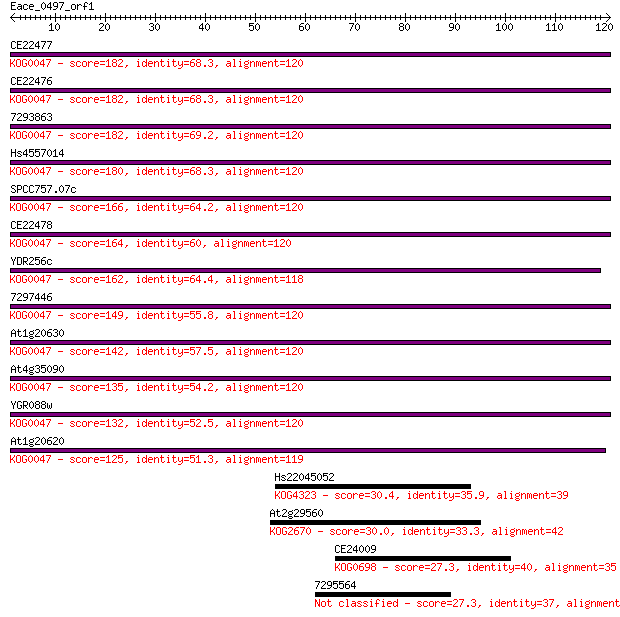
CE22477 182 1e-46
CE22476 182 1e-46
7293863 182 2e-46
Hs4557014 180 5e-46
SPCC757.07c 166 8e-42
CE22478 164 3e-41
YDR256c 162 2e-40
7297446 149 2e-36
At1g20630 142 1e-34
At4g35090 135 1e-32
YGR088w 132 2e-31
At1g20620 125 1e-29
Hs22045052 30.4 0.80
At2g29560 30.0 1.2
CE24009 27.3 6.5
7295564 27.3 7.5
> CE22477
Length=524
Score = 182 bits (463), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 82/120 (68%), Positives = 104/120 (86%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGAHGYFEVT DI++YCKADIF ++G +T + +RFSTVGGE GSADTARDPRGFAI
Sbjct 95 HAKGAGAHGYFEVTHDISKYCKADIFNKVGKQTPLLIRFSTVGGESGSADTARDPRGFAI 154
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +D+V NNTP+FF+RDP+ F +FIHTQKR+PQT+LK+ +M++ F+ PE+LH
Sbjct 155 KFYTEEGNWDLVGNNTPIFFIRDPIHFPNFIHTQKRNPQTHLKDPNMIFDFWLHRPEALH 214
> CE22476
Length=500
Score = 182 bits (463), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 82/120 (68%), Positives = 104/120 (86%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGAHGYFEVT DI++YCKADIF ++G +T + +RFSTVGGE GSADTARDPRGFAI
Sbjct 71 HAKGAGAHGYFEVTHDISKYCKADIFNKVGKQTPLLIRFSTVGGESGSADTARDPRGFAI 130
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +D+V NNTP+FF+RDP+ F +FIHTQKR+PQT+LK+ +M++ F+ PE+LH
Sbjct 131 KFYTEEGNWDLVGNNTPIFFIRDPIHFPNFIHTQKRNPQTHLKDPNMIFDFWLHRPEALH 190
> 7293863
Length=506
Score = 182 bits (461), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 83/120 (69%), Positives = 99/120 (82%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GYFEVT DITQYC A IF ++ +T + VRFSTVGGE GSADTARDPRGFA+
Sbjct 73 HAKGAGAFGYFEVTHDITQYCAAKIFDKVKKRTPLAVRFSTVGGESGSADTARDPRGFAV 132
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ +G++D+V NNTPVFF+RDP+ F FIHTQKR+PQT+LK+ DM W F +L PES H
Sbjct 133 KFYTEDGVWDLVGNNTPVFFIRDPILFPSFIHTQKRNPQTHLKDPDMFWDFLTLRPESAH 192
> Hs4557014
Length=527
Score = 180 bits (457), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 82/120 (68%), Positives = 100/120 (83%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKGAGA GYFEVT DIT+Y KA +F+ IG KT + VRFSTV GE GSADT RDPRGFA+
Sbjct 75 HAKGAGAFGYFEVTHDITKYSKAKVFEHIGKKTPIAVRFSTVAGESGSADTVRDPRGFAV 134
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ +G +D+V NNTP+FF+RDP+ F FIH+QKR+PQT+LK+ DM+W F+SL PESLH
Sbjct 135 KFYTEDGNWDLVGNNTPIFFIRDPILFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLH 194
> SPCC757.07c
Length=512
Score = 166 bits (421), Expect = 8e-42, Method: Compositional matrix adjust.
Identities = 77/120 (64%), Positives = 95/120 (79%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKG+GA G FE TDDIT+Y K +F ++G KT + RFSTVGGERG+ DTARDPRGFA+
Sbjct 60 HAKGSGAFGEFECTDDITKYTKHTMFSKVGKKTPMVARFSTVGGERGTPDTARDPRGFAL 119
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EGI+DMV NNTPVFF+RDP KF FIHTQKR+PQ ++K+ M W + S + ES+H
Sbjct 120 KFYTDEGIFDMVGNNTPVFFLRDPAKFPLFIHTQKRNPQNDMKDATMFWDYLSQNAESIH 179
> CE22478
Length=512
Score = 164 bits (416), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 72/120 (60%), Positives = 97/120 (80%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKG GAHGYFEVT DIT+YCKAD+F ++G +T + VRFSTV GE GSADT RDPRGF++
Sbjct 86 HAKGGGAHGYFEVTHDITKYCKADMFNKVGKQTPLLVRFSTVAGESGSADTVRDPRGFSL 145
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ EG +D+V NNTP+FF+RD + F +FIH KR+PQT++++ + ++ F+ PES+H
Sbjct 146 KFYTEEGNWDLVGNNTPIFFIRDAIHFPNFIHALKRNPQTHMRDPNALFDFWMNRPESIH 205
> YDR256c
Length=515
Score = 162 bits (409), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 76/118 (64%), Positives = 92/118 (77%), Gaps = 1/118 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA G+GA GYFEVTDDIT C + +F +IG +T+ RFSTVGG++GSADT RDPRGFA
Sbjct 70 HAHGSGAFGYFEVTDDITDICGSAMFSKIGKRTKCLTRFSTVGGDKGSADTVRDPRGFAT 129
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPES 118
KFY+ EG D V NNTPVFF+RDP KF FIHTQKR+PQTNL++ DM W F + +PE+
Sbjct 130 KFYTEEGNLDWVYNNTPVFFIRDPSKFPHFIHTQKRNPQTNLRDADMFWDFLT-TPEN 186
> 7297446
Length=506
Score = 149 bits (375), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 67/120 (55%), Positives = 90/120 (75%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
+AKGAGA GYFE T DI+++C A IF ++ +T V +RFS GE+GSADT R+ RGFA+
Sbjct 73 YAKGAGAFGYFECTHDISKFCAASIFDKVRKRTAVAMRFSVACGEQGSADTVREQRGFAV 132
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+ +GI+D+V N PV +VRDP+ F +H QKR+PQT+LK+ DM W F +L PE+LH
Sbjct 133 KFYTDDGIWDIVGCNMPVHYVRDPMLFPSLVHAQKRNPQTHLKDPDMFWDFMTLRPETLH 192
> At1g20630
Length=492
Score = 142 bits (359), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 69/120 (57%), Positives = 88/120 (73%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA+GA A G+FEVT DITQ AD + G++T V VRFSTV ERGS +T RDPRGFA+
Sbjct 65 HARGASAKGFFEVTHDITQLTSADFLRGPGVQTPVIVRFSTVIHERGSPETLRDPRGFAV 124
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+REG +D+V NN PVFFVRD +KF D +H K +P+++++ N + FFS PESLH
Sbjct 125 KFYTREGNFDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLH 184
> At4g35090
Length=492
Score = 135 bits (341), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 65/120 (54%), Positives = 87/120 (72%), Gaps = 0/120 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA+GA A G+FEVT DI+ AD + G++T V VRFSTV ERGS +T RDPRGFA+
Sbjct 65 HARGASAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVIHERGSPETLRDPRGFAV 124
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESLH 120
KFY+REG +D+V NN PVFF+RD +KF D +H K +P+++++ N + FFS PESL+
Sbjct 125 KFYTREGNFDLVGNNFPVFFIRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLN 184
> YGR088w
Length=573
Score = 132 bits (331), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 63/123 (51%), Positives = 85/123 (69%), Gaps = 3/123 (2%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HAKG G FE+TD ++ A ++ +G K VRFSTVGGE G+ DTARDPRG +
Sbjct 75 HAKGGGCRLEFELTDSLSDITYAAPYQNVGYKCPGLVRFSTVGGESGTPDTARDPRGVSF 134
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNL---KNNDMMWGFFSLSPE 117
KFY+ G +D V NNTPVFF+RD +KF FIH+QKRDPQ++L ++ + W + +L+PE
Sbjct 135 KFYTEWGNHDWVFNNTPVFFLRDAIKFPVFIHSQKRDPQSHLNQFQDTTIYWDYLTLNPE 194
Query 118 SLH 120
S+H
Sbjct 195 SIH 197
> At1g20620
Length=531
Score = 125 bits (315), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 61/119 (51%), Positives = 82/119 (68%), Gaps = 0/119 (0%)
Query 1 HAKGAGAHGYFEVTDDITQYCKADIFKEIGLKTEVFVRFSTVGGERGSADTARDPRGFAI 60
HA+G A G+FEVT DI+ AD + G++T V VRFSTV ER S +T RD RGFA+
Sbjct 104 HARGISAKGFFEVTHDISNLTCADFLRAPGVQTPVIVRFSTVVHERASPETMRDIRGFAV 163
Query 61 KFYSREGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQTNLKNNDMMWGFFSLSPESL 119
KFY+REG +D+V NNTPVFF+RD ++F D +H K +P+TN++ + + S PESL
Sbjct 164 KFYTREGNFDLVGNNTPVFFIRDGIQFPDVVHALKPNPKTNIQEYWRILDYMSHLPESL 222
> Hs22045052
Length=580
Score = 30.4 bits (67), Expect = 0.80, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 3/39 (7%)
Query 54 DPRGFAIKFYSREGIYDMVCNNTPVFFVRDPLKFVDFIH 92
+P F +FY + VCN P + R PL++VD +H
Sbjct 230 EPMMFGDRFYL---FFCSVCNQGPEYIERLPLRWVDVVH 265
> At2g29560
Length=475
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Query 53 RDPRGFAIKFYSREGIYDM---VCNNTPVFFVRDPLKFVDFIHTQ 94
+ P F S E + DM +CN+ P+ + DP D+ HT+
Sbjct 304 KSPNKSGQNFKSAEDMIDMYKEICNDYPIVSIEDPFDKEDWEHTK 348
> CE24009
Length=356
Score = 27.3 bits (59), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 7/35 (20%)
Query 66 EGIYDMVCNNTPVFFVRDPLKFVDFIHTQKRDPQT 100
+GI+D++ N V FVR+ L +KRDPQ+
Sbjct 228 DGIWDVMTNQEVVDFVREKL-------AEKRDPQS 255
> 7295564
Length=223
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 10/27 (37%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 62 FYSREGIYDMVCNNTPVFFVRDPLKFV 88
FY ++ IY + CN +F++ P FV
Sbjct 142 FYLKKSIYGLFCNKRFLFYLSSPATFV 168
Lambda K H
0.323 0.140 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40