bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
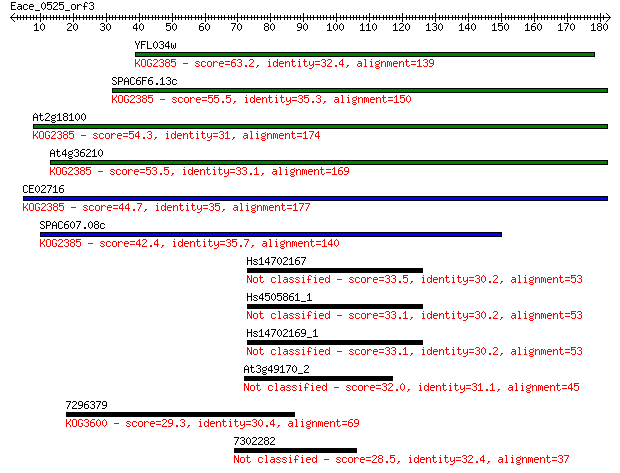
Query= Eace_0525_orf3
Length=182
Score E
Sequences producing significant alignments: (Bits) Value
YFL034w 63.2 3e-10
SPAC6F6.13c 55.5 5e-08
At2g18100 54.3 1e-07
At4g36210 53.5 2e-07
CE02716 44.7 1e-04
SPAC607.08c 42.4 5e-04
Hs14702167 33.5 0.25
Hs4505861_1 33.1 0.30
Hs14702169_1 33.1 0.32
At3g49170_2 32.0 0.62
7296379 29.3 4.5
7302282 28.5 8.0
> YFL034w
Length=1073
Score = 63.2 bits (152), Expect = 3e-10, Method: Composition-based stats.
Identities = 45/140 (32%), Positives = 75/140 (53%), Gaps = 5/140 (3%)
Query 39 GIAAAVASLGSLPLSAFLASAGGMAALVSIFGAGGAGLTGWKYSRRIANIKIFEFLMLNG 98
GIAA ++++G ++FL GG + A GA + S+R+ +++ FEF L+
Sbjct 569 GIAAGLSTIGITGATSFLTGVGGTTVVAVSSTAIGANIGARGMSKRMGSVRTFEFRPLHN 628
Query 99 RSPSSLRVGIGVSGYL-RDDDDVTLPWVCSFPNPRCDLHALKWEPHVLKALGGMVIKMVS 157
+L + VSG++ ++DDV LP+ P DL++L WEP +LK++G V + +
Sbjct 629 NRRVNLI--LTVSGWMVGNEDDVRLPFSTVDP-VEGDLYSLYWEPEMLKSIGQTVSIVAT 685
Query 158 QDFAVSASKFYLQYTVLGGL 177
+ F S + L TVL L
Sbjct 686 EIFTTSLQQI-LGATVLTAL 704
> SPAC6F6.13c
Length=778
Score = 55.5 bits (132), Expect = 5e-08, Method: Composition-based stats.
Identities = 53/158 (33%), Positives = 84/158 (53%), Gaps = 13/158 (8%)
Query 32 TAGLAAP----GIAAAVASLG--SLPLSAFLASAGGMAALVSIFGA-GGAGLTGWKYSRR 84
++GL AP GI AA ++G + S FLA GG AAL++ GA GA + + R
Sbjct 373 SSGLLAPIISAGIGAAFTTVGLSGVATSGFLA-GGGSAALITAGGAISGAHIGTTGMAHR 431
Query 85 IANIKIFEFLMLNGRSPSSLRVGIGVSGY-LRDDDDVTLPWVCSFPNPRCDLHALKWEPH 143
A++K FEF L+ + ++ V + VSG+ L +DDV L + P D++++ WEP
Sbjct 432 KADVKTFEFRPLHAQRRAN--VIVTVSGWMLSKEDDVRLSFATLDP-IVGDIYSVFWEPE 488
Query 144 VLKALGGMVIKMVSQDFAVSASKFYLQYTVLGGLTFAL 181
+L A G + +++ + + + L TVL L AL
Sbjct 489 ML-ASAGQTMNILATEVVTQSLQQVLGSTVLVSLMGAL 525
> At2g18100
Length=655
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 54/189 (28%), Positives = 87/189 (46%), Gaps = 17/189 (8%)
Query 8 SKQKTWRRLKIAGAA-VGGGVLLALTAGLAAPGIAAAVASLGS-----LPLSAFLASAGG 61
S+ W+R I GAA + GG L+A+T GLAAP IAA +L +P+ A
Sbjct 293 SEWAKWKRGGIVGAAALTGGTLMAITGGLAAPAIAAGFGALAPTLGTIVPVIGASGFAAA 352
Query 62 MAALVSIFGAGG---------AGLTGWKYSRRIANIKIFEFLMLNGRSPSSLRVGIGVSG 112
A ++ G+ AGLTG K +RR +I+ FEF + + V I V+G
Sbjct 353 AEAAGTVAGSVAVAASFGAAGAGLTGTKMARRTGDIEEFEFKAIGENHNQGVTVEILVAG 412
Query 113 YLRDDDDVTLPWVCSFPNPRCDLHALKWEPHVLKALGGMVIKMVSQDFAVSASKFYLQYT 172
++ ++D PW N + + ++WE + A+ + ++ A+ + T
Sbjct 413 FVLKEEDFVKPWEGLTSN--LERYTVQWESENIIAVSTAIQDWLTSRVAMELMRQGAMRT 470
Query 173 VLGGLTFAL 181
VL L A+
Sbjct 471 VLNSLLAAM 479
> At4g36210
Length=516
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 56/185 (30%), Positives = 87/185 (47%), Gaps = 18/185 (9%)
Query 13 WRRLKIAGAA-VGGGVLLALTAGLAAPGIAAAVASLGS-----LPLSAFLASAGGMAALV 66
W+R I GAA + GG L+A+T GLAAP IAA +L +P+ A +A
Sbjct 192 WKRGGIIGAAAITGGTLMAITGGLAAPAIAAGFGALAPTLGTIIPVIGAGGFAAAASAAG 251
Query 67 SIFGAGG---------AGLTGWKYSRRIANIKIFEFLML-NGRSPSSLRVGIGVSGYLRD 116
++ G+ AGLTG K +RRI ++ FEF + + L V + V+G + +
Sbjct 252 TVAGSVAVAASFGAAGAGLTGTKMARRIGDLDEFEFKAIGENHNQGRLAVEVLVAGVVFE 311
Query 117 DDDVTLPWVCSFPNPRCDLHALKWEPHVLKALGGMVIKMVSQDFAVSASKFYLQYTVLGG 176
++D PW N + + L+WE L + + ++ A+ K +TVL
Sbjct 312 EEDFVKPWEGLTSN--LERYTLQWESKNLILVSTAIQDWLTSRLAMELMKQGAMHTVLAS 369
Query 177 LTFAL 181
L AL
Sbjct 370 LLMAL 374
> CE02716
Length=531
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 62/180 (34%), Positives = 89/180 (49%), Gaps = 7/180 (3%)
Query 5 KEGSKQKTWRRLKIAGAAVGGGVLLALTAGLAAPGIAAAVASLGSLPLSAFLASAGGMAA 64
K +K R L I A GGVL+ LT GLAAP +AA+ L A LA+ G A
Sbjct 205 KTARNKKIKRYLMIGAAGGVGGVLIGLTGGLAAPLVAASAGMLIGGGAVAGLATTAGAAV 264
Query 65 LVSIFGAGGAGLTGWKYSRRIANIKIFEFLMLNGRSPSSLRVGIGVSGYLRDDDDVTLPW 124
L + G GAG TG+K +R+ I+ F L+ SL + VSG++ D +
Sbjct 265 LGTTMGVAGAGFTGYKMKKRVGAIEEFSVETLS--EGVSLSCSLVVSGWIESDTSPDQAF 322
Query 125 VCSFPNPRC--DLHALKWEPHVLKALGGMVIKMVSQDFAVS-ASKFYLQYTVLGGLTFAL 181
V + + R + + L++E + L LG + ++S FAVS A + L T L GL A+
Sbjct 323 VHQWRHLRHTKEQYTLRYESNYLMELGNAIEYLMS--FAVSVAIQQTLLETALAGLVSAV 380
> SPAC607.08c
Length=579
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 50/154 (32%), Positives = 76/154 (49%), Gaps = 19/154 (12%)
Query 10 QKTWRRLKIAGAAVGGGVLLALTAGLAAPGIAAAVASL----------GSLPLSAFLASA 59
+K RR+ + A + GG L+ LT GLAAP +AA + +L G+ L + SA
Sbjct 167 RKMRRRIAMGLAGLAGGALIGLTGGLAAPFVAAGLGTLFAGLGLGTMIGATYLGTLITSA 226
Query 60 GGMAALVSIFGAGGAGLTGWKYSRRIANIKIFEFLMLNGRSPSSLRVGIGVSGYLRDDDD 119
+ AL FG GA ++ + + FEF+ L+ + S L V IG+SG+L D ++
Sbjct 227 PMITAL---FGGFGAKMSMQQMGDVSKGLTDFEFIPLSVQ--SHLPVTIGISGWLGDYNE 281
Query 120 VTLPWVCSFPNPRC----DLHALKWEPHVLKALG 149
V W + D++ALK+E L LG
Sbjct 282 VDAAWKSLTVGDKSYYWGDIYALKFEVEALVDLG 315
> Hs14702167
Length=291
Score = 33.5 bits (75), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 23/53 (43%), Gaps = 7/53 (13%)
Query 73 GAGLTGWKYSRRIANIKIFEFLMLNGRSPSSLRVGIGVSGYLRDDDDVTLPWV 125
GA T W N +GR P ++R+G+G Y R+ D + PW
Sbjct 145 GAECTNW-------NSSALAQKPYSGRRPDAIRLGLGNHNYCRNPDRDSKPWC 190
> Hs4505861_1
Length=299
Score = 33.1 bits (74), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 23/53 (43%), Gaps = 7/53 (13%)
Query 73 GAGLTGWKYSRRIANIKIFEFLMLNGRSPSSLRVGIGVSGYLRDDDDVTLPWV 125
GA T W N +GR P ++R+G+G Y R+ D + PW
Sbjct 145 GAECTNW-------NSSALAQKPYSGRRPDAIRLGLGNHNYCRNPDRDSKPWC 190
> Hs14702169_1
Length=253
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 23/53 (43%), Gaps = 7/53 (13%)
Query 73 GAGLTGWKYSRRIANIKIFEFLMLNGRSPSSLRVGIGVSGYLRDDDDVTLPWV 125
GA T W N +GR P ++R+G+G Y R+ D + PW
Sbjct 99 GAECTNW-------NSSALAQKPYSGRRPDAIRLGLGNHNYCRNPDRDSKPWC 144
> At3g49170_2
Length=311
Score = 32.0 bits (71), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 72 GGAGLTGWKYSRRIANIKIFEFLMLNGRSPSSLRVGIGVSGYLRD 116
G A TG Y + ++ I E ++N +S ++ IG GY++D
Sbjct 132 GDARFTGLVYGSNLTDVVITEVTLINTQSAIRIKTAIGRGGYIKD 176
> 7296379
Length=2444
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 32/69 (46%), Gaps = 4/69 (5%)
Query 18 IAGAAVGGGVLLALTAGLAAPGIAAAVASLGSLPLSAFLASAGGMAALVSIFGAGGAGLT 77
+AGAA VL +L+ PG A+A +S+G+ P SA G + G G T
Sbjct 368 VAGAAAESAVLASLSC---PPGAASAPSSMGAAP-SATAVPLGPASLNAPALAGAGVGAT 423
Query 78 GWKYSRRIA 86
++ I+
Sbjct 424 ASSAAKEIS 432
> 7302282
Length=614
Score = 28.5 bits (62), Expect = 8.0, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 69 FGAGGAGLTGWKYSRRIANIKIFEFLMLNGRSPSSLR 105
+G GG+ L W++ R +I + + +M + RSP +R
Sbjct 334 YGLGGSALNKWRHITRDLHIDLQKGIMGDKRSPLKIR 370
Lambda K H
0.322 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2878611680
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40